I. INTRODUCTION
Coronavirus is a member of a vast family of viruses that vary from the common cold to a life-threatening infection. The first ever case of Severe Acute Respiratory Syndrome Coronavirus 1 (SARS-COV1) was reported in 2002 in Yunnan, China and spread over to 30 countries, with more than 8000 diagnosed cases and 11% fatality rate [22]. The second, the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) epidemic started in 2013 in Saudi Arabia with over 2500 confirmed cases and a 35% fatality rate [15].
However, in 2019 a novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-COV2) case originated in Wuhan, China and spread all over the world. In February 2020, WHO termed SARS-COV2 as “COVID-19” and declared a worldwide pandemic with more than 263 million confirm diagnosed cases and 5.23 million deaths [7, 8]. A brief history of the SARS-COV is illustrated in Table 1.
| Virus Name | Pandemic Year | Virus Origin | Fatality Rate |
|---|---|---|---|
| SARS-COV1 | 2002 | China | 11% |
| MERS-COV | 2013 | Saudi Arabia | 35% |
| SARS-COV2 | 2019 | China | 1.98% |
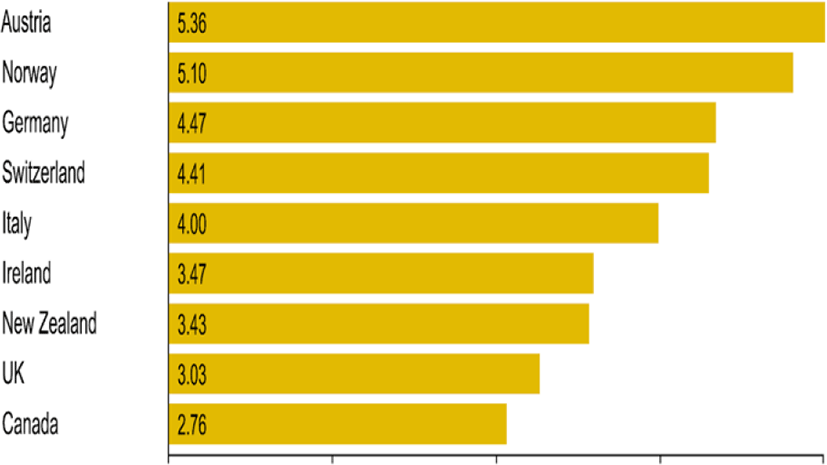
Even before the COVID-19 pandemic, the density of doctors in countries was not enough all over the world. According to data in 2020, Austria comes first with 5.36 doctors per 1000 people as shown in Figure 1. While the average in 2020 based on 9 countries was 4 doctors per 1,000 people [14].
Henceforth, the policies of nations all over the world revolve around controlling the spread of the coronavirus by implementing lockdown and making sure that the healthcare system does not collapse in the fight against COVID-19.
Multiple tests; such as Polymerase Chain Reaction (PCR), X-rays and Computed Tomography (CT), are considered as a standard test for COVID-19 diagnosis. But, PCR is very time consuming and sometimes isn’t able to detect the viral agglomeration, which results in a false negative [5].
Hence, X-rays and Computed Tomography (CT) emerged as key tests to identify the severity of the disease and its diagnosis. Sometimes they are more successful than PCR at identifying COVID 19, which might result in false negatives. Apart from the worldwide effort to develop medicine and treatment, researchers made an effort to exploit Artificial Intelligence (AI) and Deep Learning (DL) for the diagnosis of COVID-19.
In recent years, multiple AI & DL models are presented for efficient image classification. Due to this, these models are often used in Computer Aided Diagnosis (CAD) for many diseases, such as lung nodules [6, 24], heart diseases [16, 17] and brain diseases [9, 15, 22], to assist clinicians. Machine learning techniques have been designed to predict liver disease risk and outcomes like the risk of liver fibrosis, predicting liver cirrhosis, selecting a donor and survival and complications of transplant [23].
Furthermore, AI has significantly improved the overall accuracy of cancer diagnosis, medicine selection, monitoring of cancer tumors and the outcomes related to it. Researchers employed ethical and safety parameters in it to make CAD safer and build the trust of patients and clinicians. [1, 14]. However, in the field of obstetric and gynaecological, ultrasonography are two of the most used imaging procedures, AI has had little influence on this sector thus far [24].
The main contribution of this paper is as follows:
-
A lightweight sequential classification model has been presented to classify and diagnose COVID-19 patients using X-ray images.
-
This study has been performed on a very extensive dataset of more than 22 thousand X-ray Images.
-
This study is carried out on a binary class dataset i.e COVID-19 infected and Normal Lung X-ray image.
-
The performance of COVID-19 infected and normal lung detection using the proposed model is thoroughly evaluated on multiple variables.
-
An efficient, robust, and high accuracy base system is presented to clinicians for COVID-19 patient diagnosis.
The rest of the paper structure is as follows. Section 2 provides an overview of the current state of art proposed frameworks. Section 3 explains the methodology, dataset and proposed system architecture. Experimental results, performance of the proposed model are evaluated and discussed in Section 4. Finally, a conclusion with future directions is discussed in Section 5.
II. RELATED WORK
In this section, we will discuss some state-of-the-art framework of CNN and its widespread application.
In [4], the authors presented a framework to efficiently detect the fight detection using blur information by employing CNN and transform learning. The proposed scheme accurately detects 56% and 75% fights in hockey video.
Face detection is used many fields such as security, expression detection and person identification. In [19], authors presented a novel framework of frontal face generation from multi-view face images. using Generative Adversarial Network (GAN). The proposed scheme considers various factors such as beard, ear, nose and mouth. While the authors of [16], presented a facial expression detection and identification by geometric and feature extraction information and employing CNN for better results. The proposed scheme has achieved an accuracy of 96.5% on EXTENDED COHN-KANADE (CK+) dataset. Moreover, it has achieved 91.3% accuracy to detect facial expression on JAFFE dataset.
The researchers in [3] presented a DL model to explore current and commercial medications for “drug repositioning”, which is the process of designing a fast treatment approach by utilizing existing medicines that may be given to infected patients promptly. Because it often takes years for new medicines to be successfully evaluated and approved before they are released to the market.
Singh et al. [10] presented a Convolutional Neural Network (CNN) based multi-objective differential evolution (MODE) framework to efficiently classify chest computed tomography (CT) images from persons infected with and without COVID-19.
The authors of [14] presented a model to improve the Visual Geometry Group (VGG19) and the Google Mobile Network’s performance. The author used a dataset of 50 X-ray imaging, 25 of which were COVID-19 positive. The proposed approach has a 90 % overall accuracy for identifying patients.
X. Chen et al. [29] presented an automated framework; Residual Attention U-Net, to classify and diagnose COVID-19 patients using CT images. They investigate the COVID-19 link with pneumonia and its effect on the lungs. Adhikari [21] proposed a framework called “Auto Diagnostic Medical Analysis (ADMA)” which searches and identifies infectious parts of lungs to assist clinicians. In their research, they have used X-ray and CT imaging for efficient diagnosis. In addition to that, the DenseNet network can be opted to eliminate and label infectious regions. Shuai, et al. [26] proposed a hybrid transfer learning model with multi-validation (internal and external) to diagnose the COVID-19 patient. In their paper, they only used CT scan images for classification and achieved an overall accuracy of 89.5% with a sensitivity of 87%.
Narin et al. [2] proposed a ResNet50, Inception v3, and Inception-ResNetV2 base hybrid DL model to detect the COVID-19 infected lung through X-ray imaging. Goshal et al. [11] presented a Bayesian CNN and VGG16 based framework to diagnose COVID-19 using X-ray images. They improved the cognition of DL to accurately and efficiently diagnose the COVID-19 patient.
A comparison analysis of all the related work papers in terms of accuracy and precision is illustrated in Table 2.
| Sr. | Modality | Datasets Samples | Number of Classes | Results | |
|---|---|---|---|---|---|
| Accuracy | Precision | ||||
| [2] | X-ray | 8088 | 4 | 96.1% | 76.5% |
| [10] | CT scan | 40 | 2 | 93.4% | N/A |
| [14] | X-ray | 50 | 2 | 90% | 91.5% |
| [21] | X-ray and CT scan | 4344 | 2 | 89% | 90.1% |
| [26] | CT scan | 1065 | 3 | 89.5% | N/A |
| [29] | CT scan | 110 | 2 | 89% | 95% |
III. METHODOLOGY
In this section, we explain the dataset, system architecture, proposed framework, and experimental environment.
Dataset plays a very pivotal role in generating better accuracy of the system because with a bigger dataset the system can be trained more thoroughly. In addition to that, results that are acquired from systems that are trained on a very scarce dataset can never be trusted or have a very little confidence in.
In this study, a dense chest X-ray images dataset has been provided by Dr. Muhammad Haziq Khan1. The acquired dataset consists of mainly SARS-COV2 patients and normal person chest X-rays images.
Overall, the dataset contains a total of 22,294 image samples, out of which 6276 are COVID-19 patients while the rest are healthy chest X-rays. The collected dataset samples range from 1112 x 624 to 2170 x 1953 pixels. The dataset is randomly divided into training and testing datasets with a 70% to 30% ratio respectively.
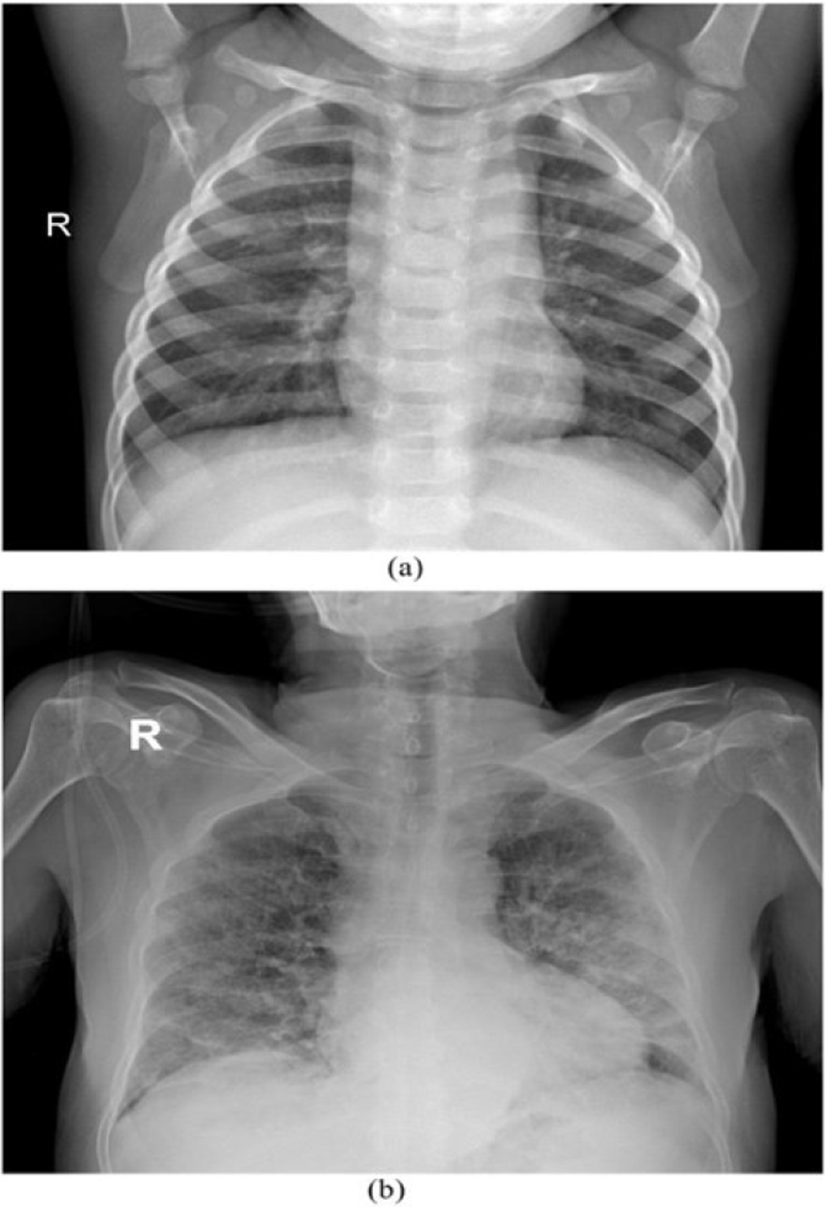
Our experimentation is based on the binary class dataset i.e., COVID-19 infected and normal Lung X-ray image as show in Figure 2.
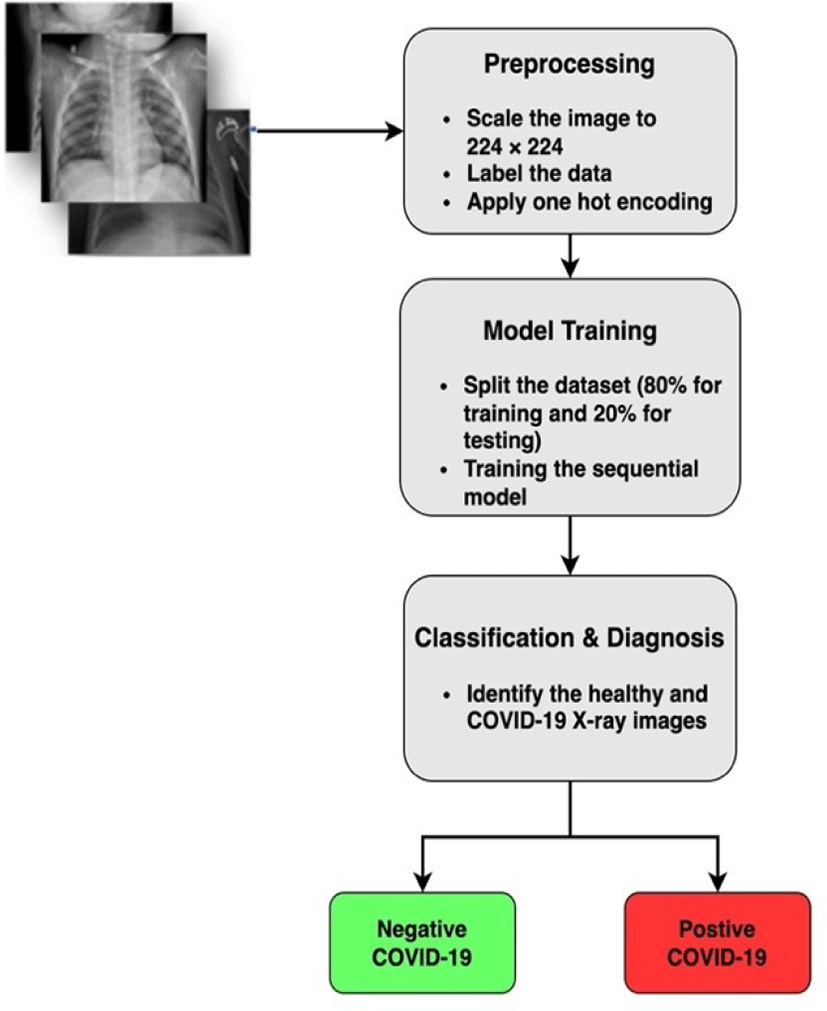
We presented a lightweight CNN model for automatic and efficient COVID-19 image classification. Fig 3 displays the overall working of the proposed system. The proposed system can be divided into three parts.
The dataset is very diverse in terms of sizes ranging from 1112 x 624 to 2170 x 1953 pixels. To create uniformity and consistency, we rescale the sample images into 224 x 224 pixels. Furthermore, one hot encoding is applied based on labels of data to create a distinction between the positive and negative COVID-19 X-ray image.
Before training we split the dataset into 2 parts (Training and Testing ) with a 70 to 30 ratio. This means that out of 22,294 X-ray samples we use 30% (6687 samples) for testing while using 70% (15607 samples) for the training phase.
In addition to that image segmentation and feature extraction is implemented and those features are forwarded to the proposed lightweight sequential model for training.
After the training phase is completed, testing data samples are forwarded to the trained model for evaluation. The trained model automatically classifies the dataset into COVID-19 positive or negative as shown in Figure 3. Furthermore, the performance of the overall system is evaluated based on multiple parameters such as F1-score, accuracy, specificity etc.
We implemented the model in Keras on Tensorflow v2 with a learning rate of 0.01 using Adam optimizer and a total 25 number of epochs were set for model training. The experimentation was performed on the Jupyter Notebook equipped with GeForce RTX 2080 Ti GPU. The main code is written in python, related version details are shown in Table 3.
| Experimental Setup | Version |
|---|---|
| CUDA | 11.2 |
| Operating System | Ubuntu 18.04 |
| GPU | GeForce RTX 2080 Ti |
| Python | 3.6.9 |
Each image sample is transformed into a greyscale. Furthermore, to minimize the biases of the model training, we used shear image, shuffling, image rotation and flip, zoom and rescaling. We constitute that shear range (0.2), zooming (0.3) and horizontal flip are most apt for training.
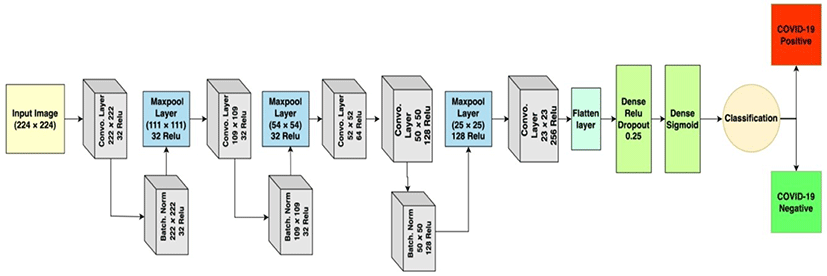
The implemented model has a multi convolutional layer, each of which is followed by max-pooling and batch normalization. A derail structure of the model in block diagram is illustrated in Figure 4.
IV. RESULT AND DISCUSSION
The model has been thoroughly evaluated on multi-performance parameters i.e., accuracy, precision, specificity etc. Furthermore, we train and tested our proposed model on publicly 2 available datasets to validate its performance.
Confusion Matrix is a salient method to analyze the performance of a model. It comprises four parameters True Positive (TP), False Positive (FP), False Negative (FN), and True Negative (TN).
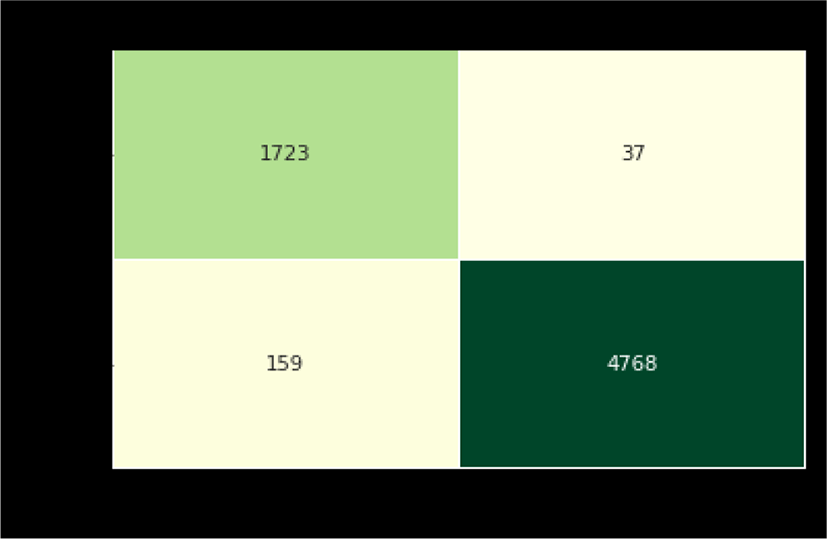
We evaluate our proposed model on binary class dataset samples (COVID-19 and Normal X-ray images). A total of 6687 samples are selected for testing containing 1760 COVID-19 and 4927 healthy X-ray images respectively. From the confusion matrix (as shown in the Figure 5), it is clear that we have successfully diagnosed 1723 out of 1760 COVID-19 patients, thus achieving 97.38% accuracy. At the same time, we have successfully detected 4768 normal people out of 4927 and achieved an accuracy of 96.77%.
This indicates that the presented model has achieved an overall accuracy of 96.88% to diagnose COVID-19 patients successfully from the randomly chosen dataset. In addition to that, a detailed analysis of the results has been presented in Table 4.
| Class | Precision | Recall | F1 Score |
|---|---|---|---|
| Overall | 95.5% | 97.5% | 96.5% |
| COVID-19 | 92% | 98% | 95% |
| Normal | 99% | 97% | 98% |
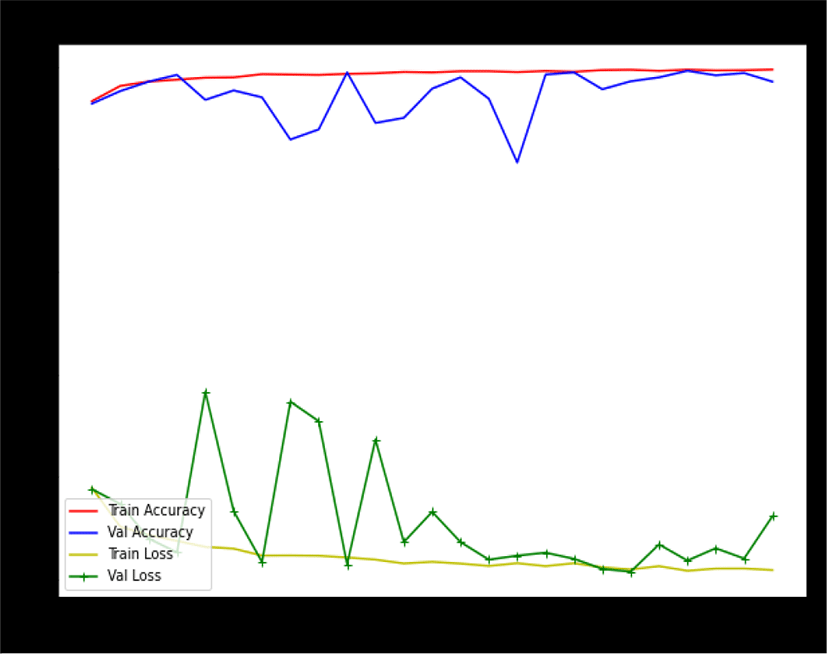
We train our model for 25 epochs with a learning rate of 0.01. The average training accuracy is more than 94% with an error rate of 0.3%. However, during the training of the model, it is noted that training accuracy of more than 98% accuracy with a 0.012% error rate has been achieved (as shown in Figure 6) which is quite good. Such accuracy gives strengths and confidence in the model, especially in the CAD system.
The proposed model has been thoroughly validated by using publicly available Chest X-ray datasets2. The dataset2 [11] contains both CT scan and chest x-ray images. However, we only use X-ray images to validate and calculate the accuracy of model on publicly available dataset2. It has a total of 9546 (5501 Normal and 4045 COVID-19) X-ray images.
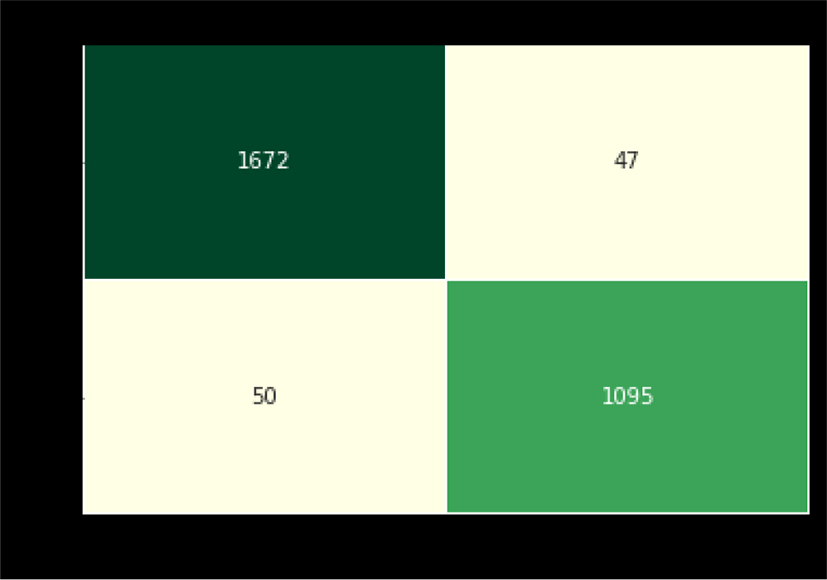
A total of 6682 images are randomly selected for training while 2864 images are selected for testing purposes. Among 2864 testing X-ray samples, there are 1145 healthy X-ray and 1719 COVID-19 images respectively. From Figure 7, it is clear that we have successfully diagnosed 1672 out of 1719 COVID-19 patients, thus achieving 97.26% accuracy.
While on the other hand, model has successfully detected the 1095 healthy people out of 1145 random X-ray samples. Thus, achieving an accuracy of 95.63%. In terms of an overall accuracy the proposed model has achieved an accuracy of 96.61% to efficiently make a diagnosis.
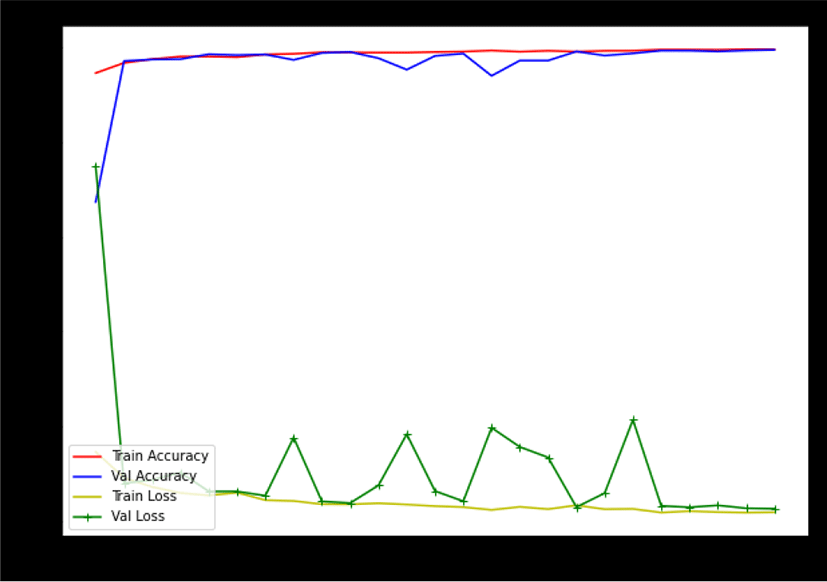
During the training of the model on the dataset2, the model have achieved an accuracy of more than 98% with an error of 0.061%. However, in some epochs the model has achieved an accuracy of more than 99% with an error rate of 0.017% as shown in Figure 8.
Although we have trained and evaluate our model on an extensive dataset, but the model still misclassifies some COVID-19 positive patients. This is wrong diagnosis happen due to hazy line of demarcation, shadow or blur effects wrong X-ray image angle (as shown in Figure 9. Furthermore, there should be a balance between both classes (COVID & Normal X-ray images) so that bias and variance between the sub-dataset (test & train) can be reduced.
V. CONCLUSION
An early diagnosis of COVID-19 is key to stopping the spread of disease to other people. Recently multiple frameworks are presented to diagnose COVID-19 by exploiting AI & DL models. However, most of these frameworks are either trained on a very scarce dataset or take too much time. In this paper, we propose a lightweight CNN model which is trained on more than 20,000 dataset samples. The proposed model has achieved an overall accuracy of 96.88% with a sensitivity of 91.55%.