I. INTRODUCTION
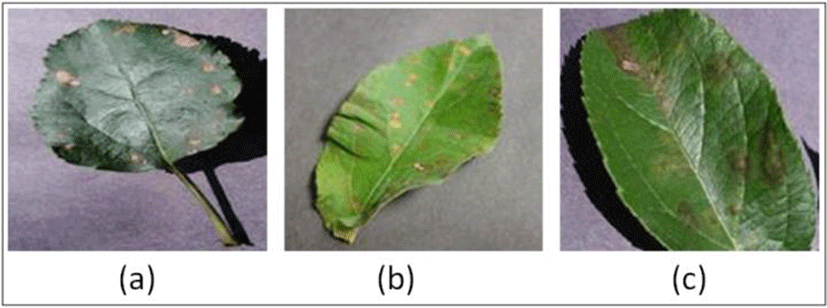
Diseases in plants can lead to a significant decrease in production and also affect the quality of agricultural products. Therefore, early identification of such diseases in plants is of utmost importance so that preventive measures can be taken in advance [16]. There are numerous symptoms of plant diseases in which mostly found in their leaves. Few examples of the infected apple leaves are depicted in Fig. 1 that are suffered from three different diseases, namely, black rot, cedar rust and scab. Traditionally, there is a requirement to diagnose such disease in plants. However, the availability of experts is limited and other factors like, proper training, assessment methodology and laboratories are required to experts for effective identification of diseases [1]. Therefore, automatic detection of plant disease has been proposed by researchers using machine learning and image processing techniques. Sena et al. [17] have proposed a methodology for the identification of damaged maize plants by the fall armyworm using digital color images. The authors subdivided the images into blocks and classified into two classes, namely, damaged or non-damaged based on the number of objects found in each block. However, their methodology was applied on a small dataset of 720 images. The title of each paper is expected to cover the whole contents of the paper and it should as concise as possible.
Disease identification in cotton corps using machine vision system has been proposed in [2]. The authors have segmented the diseased region and features that characterize the shape and appearance of an image including gray levels, connectivity and texture have been extracted. The identification of the disease has been performed using Support Vector Machine (SVM) classifier with an accuracy of 90%. The authors conclude that texture based features can be used as discriminators in absence of color or shape patters. Similarly, the authors in [3] have used Gabor wavelet filter to extract the text features from the plant leaves to improve the overall accuracy of their system. In [5], the authors have proposed the identification of citrus greening disease. High resolution aerial images have been taken using a remote sensing multi-rotor unmanned aerial vehicle (UAV). Regression analysis has been used to extract features from the acquired images where an accuracy between the range of 67% to 85% has been recorded using SVM classifier.
Advancement in deep learning techniques have led to a significant increase in solving many problems effectively [7,8,9,4]. In traditional machine-vision methods, features are extracted manually by various algorithms whereas deep learning models automatically discover higher level features from the data [10]. These networks are able to overcome the obstacles associated with traditional methods by utilizing huge amount of data and exploitation of parallel architectures. For the training of deep learning model from scratch requires the optimization of millions of parameters and a huge amount of dataset with high computing resources. Therefore, researchers move on to the technique of transfer learning, in which model is not trained from scratch rather adapt a pre-trained model by means of fine tuning with our data [23,22,21]. In this way, we utilize the knowledge learned from another similar problem to our problem [20,13]. The technique of transfer learning could be applied by two ways with the requirement that the data must match the input size of the pre-trained model. Ghazia et al. [6] have proposed plant identification using deep neural networks with the help of transfer learning technique. The authors have used three deep learning architectures, namely, GoogLeNet [19], AlexNet [9], and VGGNet [18]. They have recorded an accuracy of 80% on validation set when tested for 1000 species of plants. Similarly, the authors in [14] have used deep CNNs for plant identification. In [12], the authors have utilized a visualization technique that is based on the deconvolutional networks (DN) to gain the intuition of CNN features in plant identification using leaves. The methodology was applied on 44 plant species where an accuracy of 99.6% has been recorded. It has been found that venation structure is an important feature to identify different plant species. In this paper, we propose an assistive plant disease identification system using deep learning technique. The performance of the system has been evaluated using fine-tuning of pre-trained models such as batch size, iteration size, and data augmentation etc. For this, a popular deep learning model GoogLeNet inception architecture [19] has been used in this work to identify the disease types in the plant leaves. Rest of the paper is organized as follows. We present the details of the proposed system including designing of the feature vector and the description of the classification scheme, in Section 2. Results are discussed in Section 3. Finally, we conclude in Section 4 by discussing some future possibilities of the current work.
II. PROPOSED METHODOLOGY
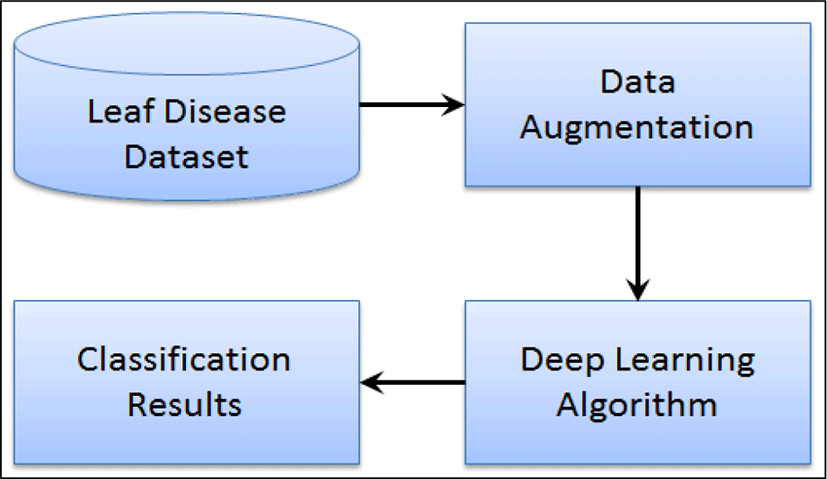
The proposed approach is based on deep learning techniques to identify the well-known diseases in apple plants leaves. The general flow diagram of the approach is depicted in Fig. 2, where image pre-processing techniques has been applied for data augmentation. Next, a pre-trained deep learning model of GoogLeNet is utilized for identification of disease types in leaves. The parameters of the pre-trained network are fine-tuned using transfer learning approach.
The image dataset of plant leaves consists of both healthy and infected leaves. All are color images having three channels i.e. Red, Green and Blue. In preprocessing step we have performed real time data augmentation where images are generated with various geometrical transformations such as horizontal flip, vertical flip, rotation, scaling, skewing, brightness [6].
In the last few years, recognition and classification of objects in images have been improved due to the introduction of deep learning and convolutional networks [11]. CNNs based architectures arise from the basic neural networks in machine learning and are proved to be very effective in recognition and classification of images. CNN’s have been successfully exploited for the recognition of MNIST digits, faces, objects, traffic signs, etc. Therefore, it has become necessary to opt for deep learning methods for new problems. Here, we have used the popular deep learning model GoogleNet for our proposed task.
GoogLeNet is an inception architecture [19] that combines the multi-scale idea and dimension reduction layers based on the Hebbian principle and embedding learning. This enables one to increase the width and depth of the network for an improved generalization capacity per a constant computational complexity. Moreover, GoogLeNet was the winner of ILSVRC 2014 [15] and consists of about 6.8 million parameters. It has nine inception modules, two convolutional layers, one convolutional layer for dimension reduction, two normalization layers, four maxpooling layers, one average pooling, one fully-connected layer, and a linear layer with Softmax activation in the output. Each inception module contains two convolutional layers, four convolutional layers for dimension reduction, and one max-pooling layer. It also uses dropout regularization in the fully-connected layer and applies the ReLU activation function in all of the convolutional layers. During training, GoogLeNet connects two auxiliary classifiers to the intermediate layers of the network to effectively perform backpropagation through all the layers. In other words, this network adds auxiliary losses to the total loss to make the earlier stages more discriminative and increase the backpropagated gradient. The additional classifiers include one average pooling layer, one convolutional layer for dimension reduction, two fully-connected layers, and a linear layer with Softmax activation.
III. EXPERIMENTAL RESULTS
Here, we present the details of the dataset used in our proposed system. The classification of diseases in leaves of apples are performed using image processing techniques with deep learning models. The details are as follows.
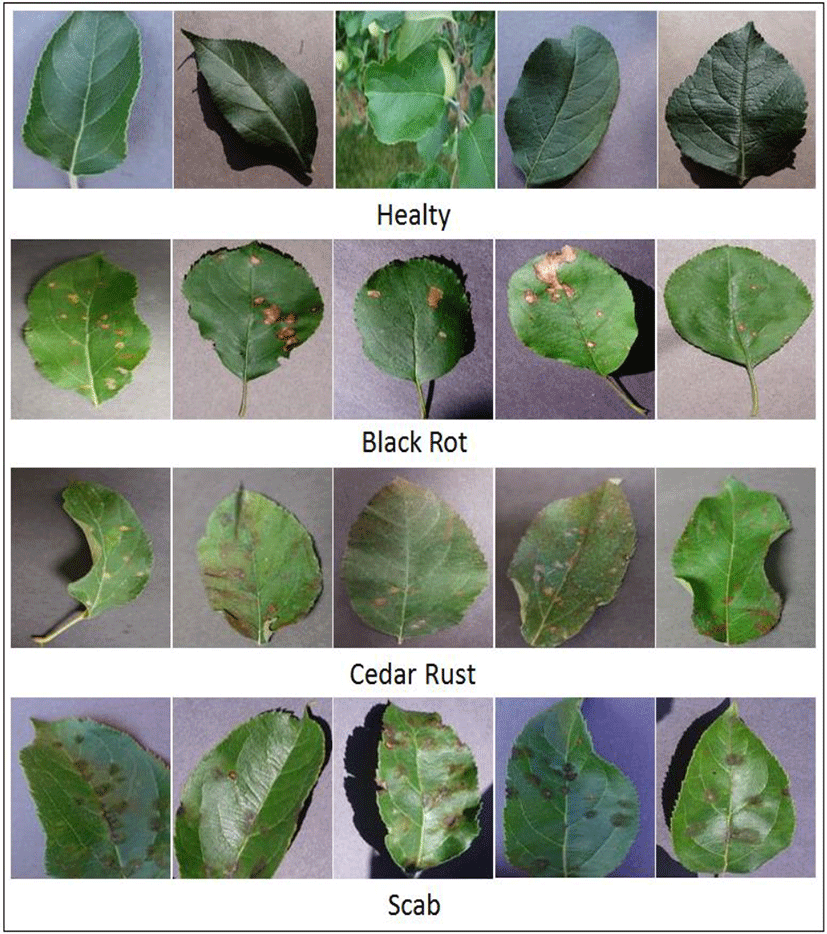
In this work, we have used the publicly available dataset consisting of color images of leaves of apple from PlantVillage dataset. The dataset consists of four class where three class represents diseases, namely, Black rot, Cedar rust and Scab whereas the fourth one is normal healthy class. The number of images in Black rot, Cedar rust, Scab and Healthy are 712, 276, 630 and 1835, respectively. Thus, a total of 3453 images of apple leaves have been analyzed in this work. The dataset has been divided into three parts i.e. training, validation and test sets as 70%, 10% and 20%, respectively. A pictorial representation of leaves of all four classes involved in this study, are depicted in Figure 3.
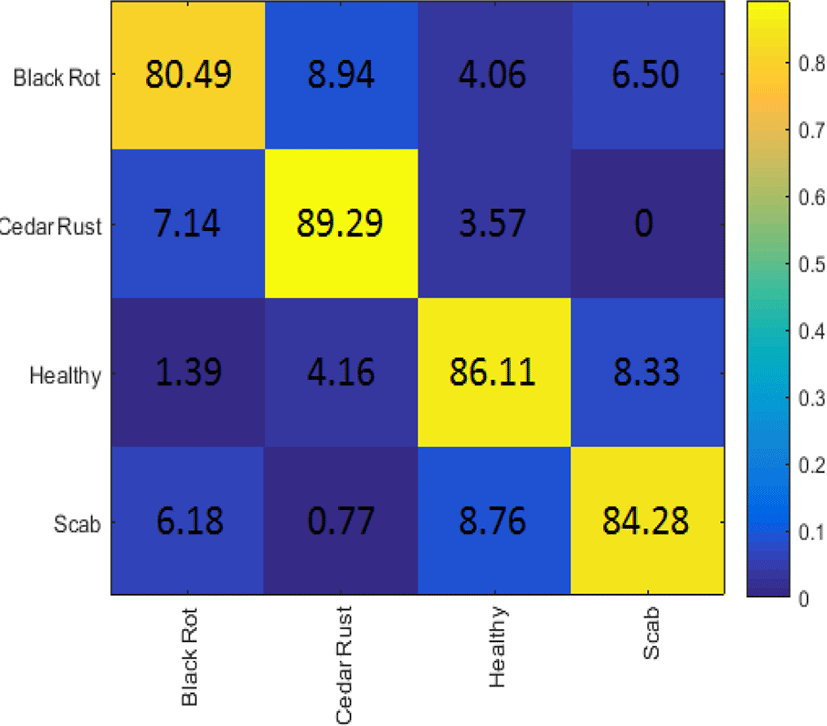
The identification of the disease was performed using transfer learning of pre-trained GoogLeNet model. Categorical Cross Entropy was used as a loss function with a learning rate of 1e-4. Four thousand epochs were run in training the network with a batch size of 100. An average accuracy of 85.04% was recorded on the four classes of the leaves. The confusion matrix of all classes is depicted in Fig. 4, where maximum and minimum accuracies of 89.29% and 80.49% were recorded in Cedar rust and Black rot classes, respectively.
IV. CONCLUSION
In this paper, we have proposed a novel assistive framework for plant disease identification that helps in early detection of diseases so that proper treatment can be made for the recognized disease. Deep learning based architecture has been utilized for the effective identification of the disease. The methodology has been applied to the disease presents in the leaves of apple plants. An average accuracy of 85.04% has been recorded using GoogLeNet model. In future, the methodology can successfully be applied to more number of diseases that can be span to large number plant species.
V. FUTURE WORKS
This paper has given details on how CNNs can be used to classify plant leaves with various diseases. Conv-Nets are not only improving for tasks like whole-image classification but also making significant progress on localization tasks which include improvements in bounding box object detection, image masking and pixel-pixel localization. Fully Convolutional Networks have shown remarkable results in pixel-pixel localization and semantic segmentation. We intend to extend the use CNNs in plant disease localization tasks later to help one study symptoms in a much better and accurate manner.