I. INTRODUCTION
India has one of the highest rates in oral cancer in the world partly attributed to high prevalence of tobacco chewing. In parallel to the increase in Oral Cancer, border-line malignant lesions which range from epithelial dysplasia to intra epithelial carcinoma have also increased in numbers [1]. In the past few years deep learning has proven to be efficient in many classification problems especially in medical imaging data. In the last few years a large number of machine learning based frameworks especially deep learning were employed by researchers to detect and classify the oral cancer. These deep learning based systems have proved to be very efficient and has achieved accuracy almost equal to the specialist pathologist that does its work manually and has ample of experience in his field [2].
The term dysplasia was introduced by Reagon in 1958 in a study where he described the features of dysplasia [3]. In medical terms dysplasia means an abnormal development in cells while histomorphologically any cellular or structural change in epithelium is dysplasia. Major pathological microscopic changes at cellular level as well as tissue level in dysplastic tissue are given in Table 1: [4].
Dysplasia is a part of the pathway to malignancy. Dysplastic cells cannot be called malignant until and unless it invades the connective tissue or metastasize. The dysplastic cells can be classified into mild, moderate and severe dysplasia.
The progressive change in dysplastic cells from mild to moderate and then from moderate to severe can be seen in the epithelium. The invasive activity of dysplastic cells starts from the lower layer, then progress to the middle layer and finally full thickness of epithelium. Next, it will invade the basement membrane [5].
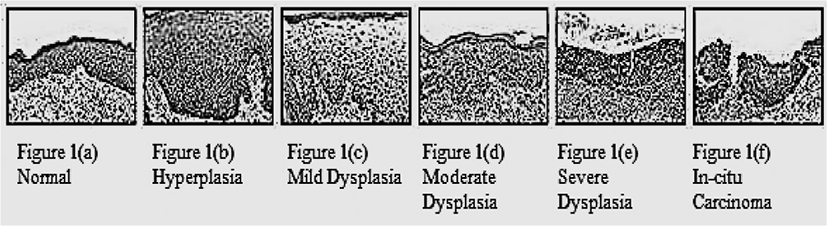
As the dysplasia arises in the basal layer of the epithelium and extends, with progression, to the upper epithelial layers, the scheme classifies mild dysplasia as involvement of the lower third of the epithelium only, moderate dysplasia as extension to the middle third, and severe dysplasia as extension to the superficial third of the epithelium [6].
Early detection plays a key role in cancer diagnosis and can improve long-term survival rates. Medical imaging is a very important technique for early cancer detection and diagnosis. As is well known, medical imaging has been widely employed for early cancer detection, monitoring, and follow-up after the treatments [7].
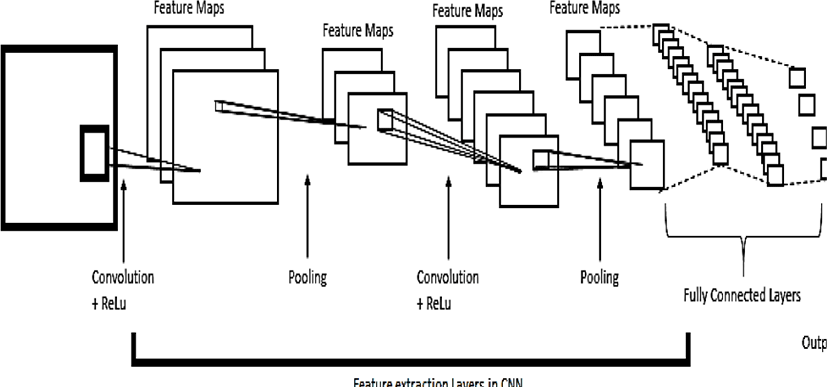
This paper aims to classify oral dysplasia by using popular deep learning technique namely Convolutional Neural Network. Since our data is in image format that’s why CNN (Convolutional Neural Network) is preferable over other deep learning techniques. Recent studies show that CNN achieve promise performance in cancer detection and diagnosis. [8, 9].
II. LITERATURE REVIEW
This section is intended to provide a brief review of recent studies on applying deep learning for early cancer detection, cancer diagnosis and prognosis.
In recent years, a bunch of papers has been published about the application of deep learning in cancer detection and diagnosis. In [10], Gustav Forslid et al. have used CNN on oral cancer dataset. Two different CNN models were used namely ResNet and VGG. Results were very good as compared to other image diagnosis techniques. In [11], Geert Litjens et al. have presented Prostate cancer identification in biopsy specimens and breast cancer metastasis detection in sentinel lymph nodes using deep learning and concluded that deep learning improves the accuracy of prostate cancer diagnosis and breast cancer staging. Authors employed CNN. In [12], Albayrak et al. developed a deep learning based feature extraction algorithm to detect mitosis in breast histopathological images. In the proposed algorithm, the CNN model was used to extract features which were used to train a support vector machine (SVM) for the detection of mitosis. In [13], Krizhevsky et al. used AlexNet to construct a CNN model to classify benign or malignant tumors from the breast histopathological images. In [14], Akshay Iyer et al. have used the pre-trained model, VGG19 to extract information from the pathological images specific for lung cancer. A model consisting of deep convolution network-based image classification has been proposed for predictions on mutations in genes signature information of lung cancer among Indian populations. In [15], Bassma El-Sherbiny et al. propose - Brain/Lung/Breast (BLB) automated detection system. It precisely predicts the occurrence of cancer and segments the expected region of tumor/cancer in MRI/CT scan/Mammography images. This system proposes different classification techniques including Support Vector Machine (SVM), ExtraTrees and convolutional neural network (CNN). CNN performed exceptionally well in the detection of lung cancer. In [16], Chen et al. proposed a deep cascade network for mitosis detection in breast histology slides. They first trained a fully connected network model to extract mitosis candidates from the whole histology slides and then fine-tuned a CaffeNet model for the classification of mitosis. Three networks with different configurations of fully connected layers were trained and the scores were averaged to generate the final output. In [17], Ida Arvidsson et al. compared two different techniques; by training the networks using color augmentation and by using digital stain separation using an auto encoder. The author achieved accuracies of 95% for classification of benign versus malignant tissue and 81% for Gleason grading for data from the same site as the training data. In [18], Albarqouni et al. explored deep CNN in a biomedical context, a multiscale CNN architecture was developed with an aggregation layer was introduced after the softmax layer to aggregate the prediction results with the annotation results from multiple participation. In [19], Sawon Pratiheret al. have deployed deep learning framework to classify elastic scattering spectra of biological tissues into normal and cancerous ones. Authors experimented to show the superiority of the convolutional neural network extracted deep features over classical handcrafted biomarkers. The proposed method employs elastic scattering spectra of the tissues as input to CNN. In [20], Wichakam et al. proposed a combined system consisting of CNN and SVM for mass detection on digital mammograms. CNN was used on mammographic patches to get the high-level feature representation of the image. This high-level feature set was used as input to SVM for classification of mammograms. In [21], Xu et al. proposed a stacked sparse auto-encoder based algorithm to classify nuclei in breast cancer histopathology. In [22], Tian Xia et al. proposed a system of tumor classification by pre-training a CNN from samples of different tissue types in histopathological images, then fine-tuning the obtained pre-trained on a particular tissue type. CNN showed improvement over training from scratch with limited data. In [23], Mina Khoshdeli et al. applied convolutional neural networks for grading of the tumor and decomposing tumor architecture from H&E stained histology sections of the kidney. In [24], Xu Y et al. proposed deep convolutional neural network activation features to perform classification, segmentation and visualization in large-scale tissue histopathology images. Authors used a pre-trained ImageNet network on features extracted by CNN.
III. MATERIAL AND METHODOLOGY
In this study we have taken data of 52 patients suffering from oral dysplasia. 2 H&E stained histopathological samples are taken from each patient. The data is taken from Indira Gandhi Govt. Dental College and Hospital, Jammu, India. Based on CIN (Cervical intraepithelial neoplasia) classification, these data samples were classified into 4 different classes namely normal, mild dysplasia, moderate dysplasia and severe dysplasia.
There are a total of 2688 images in our dataset. 663 images belonging to a normal class, 681 belonging to mild dysplasia, 675 belonging to moderate dysplasia and 669 belonging to severe dysplasia.
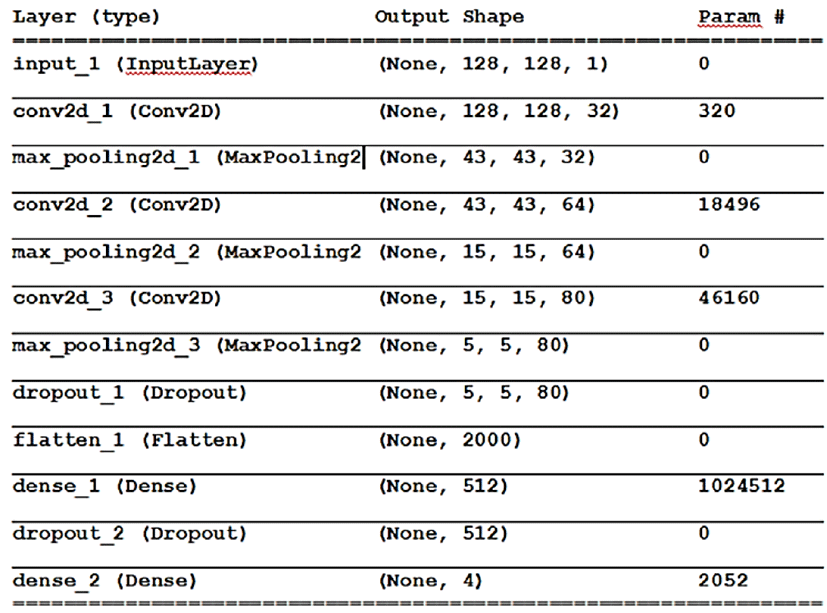
CNN was created and trained from scratch. Python language along with the Tensor flow and Keras deep learning libraries were used to build the CNN model.
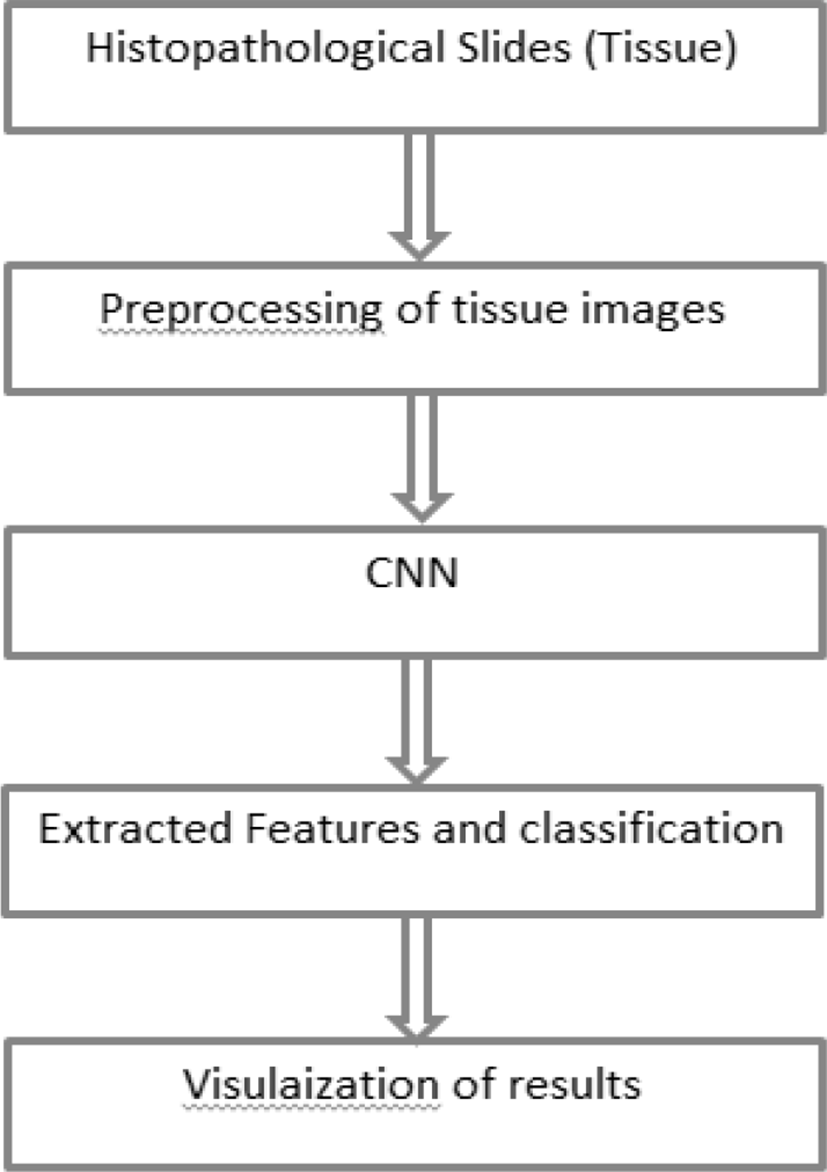
Dataset was divided into training set and testing set in a 70:30 ratio respectively. The training set contained 1882 images and testing set contained 806 images. The research methodology diagram is given in figure 4:
IV. RESULTS AND DISCUSSION
The classification of the dysplastic tissue was carried out by CNN into 4 different classes.
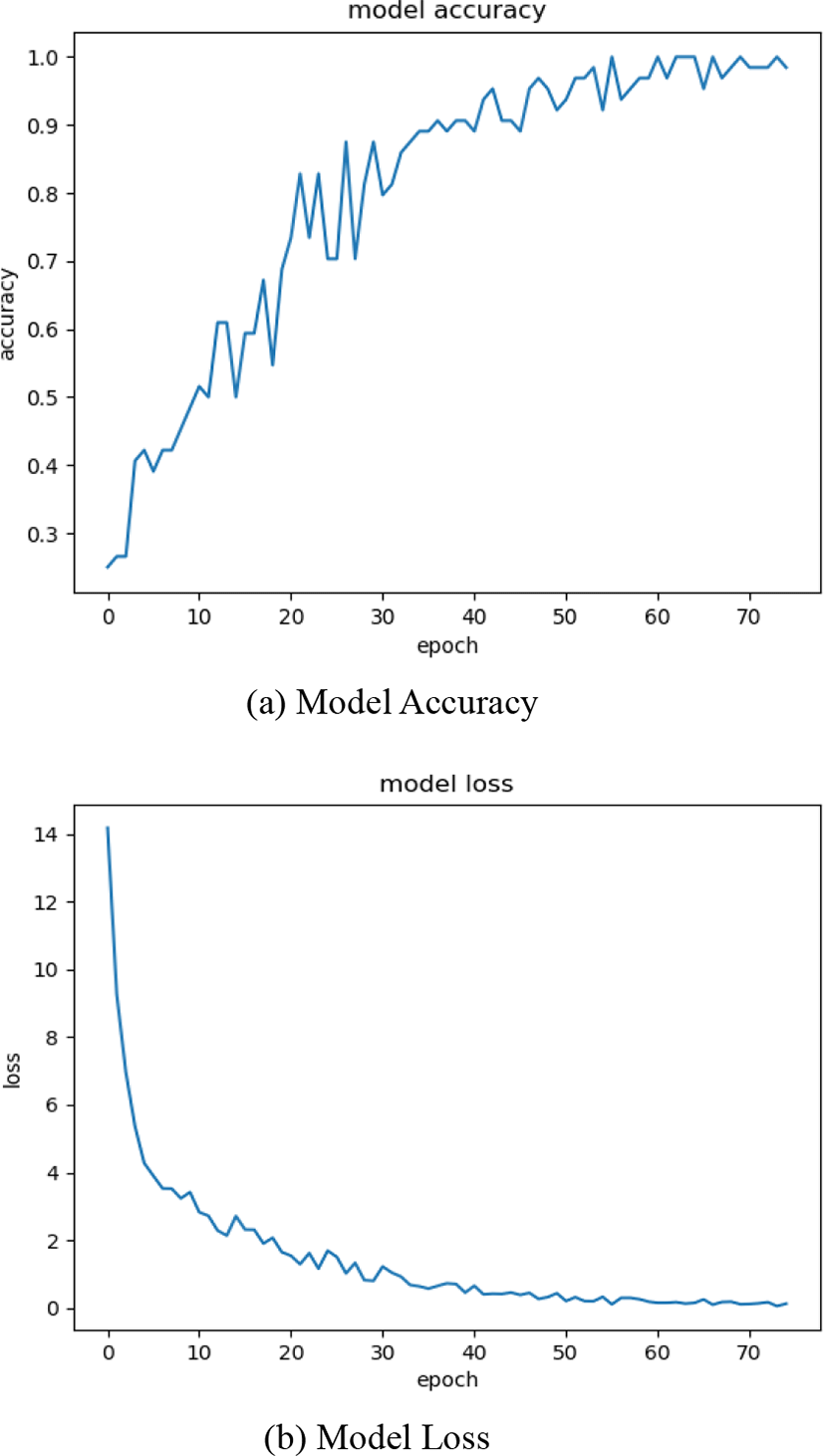
When CNN was trained on training data it gave an accuracy of 91.65% and in testing, it gave an accuracy of 89.3%. Our CNN was trained for a total of 75 epochs. The Accuracy and loss of model is given in figure 5(a) and figure 5(b) respectively.
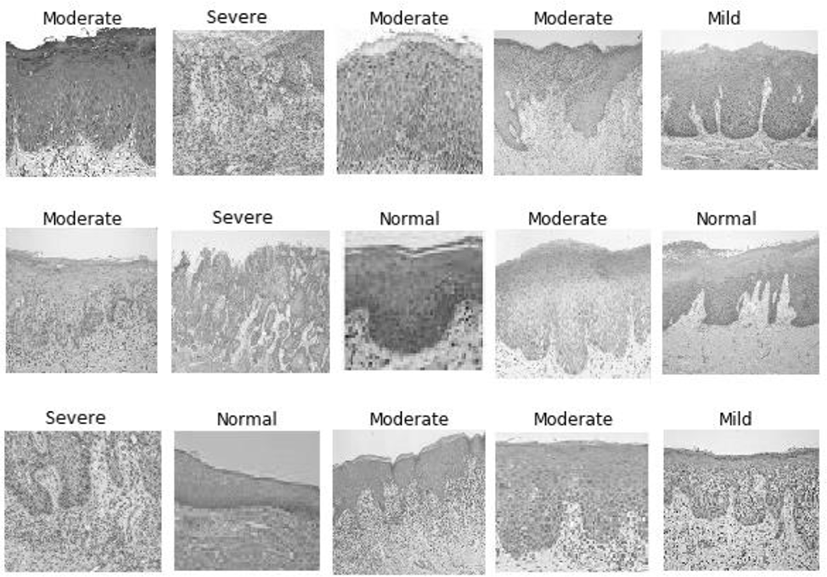
Our proposed CNN produced the following output of classified tissue images along with labels:
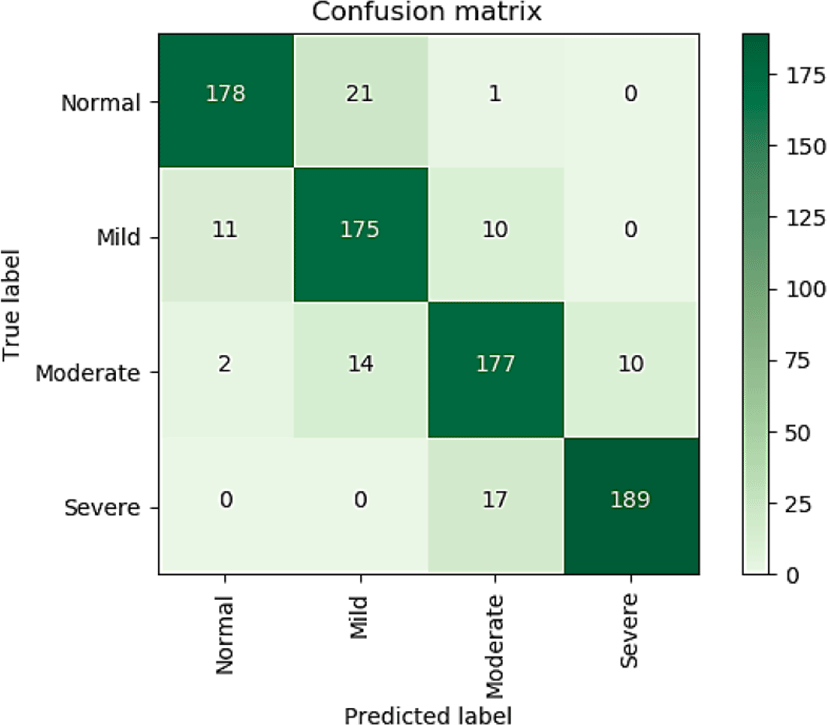
The confusion matrix obtained from the experiment is given in figure 3:
Precision, recall, f1-score for each class and average of all classes are given in Table 2.
| Class | Precision | Recall | F1-score |
|---|---|---|---|
| Normal | 0.93 | 0.89 | 0.91 |
| Mild | 0.83 | 0.89 | 0.86 |
| Moderate | 0.86 | 0.87 | 0.87 |
| Severe | 0.95 | 0.92 | 0.93 |
| Average | 0.89 | 0.89 | 0.89 |