I. INTRODUCTION
According to a report by National Cancer Registry Programme, India the cancer rates in India is increasing day by day. According to the report the number of active oral cancer patients in India is 135,929 which is second most after breast cancer. In India every year about 77,000 new cases of oral cancer are reported and about 52,000 deaths are reported due to this deadly disease [1]. Oral cancer is a cancer with high mortality rate because of the fact that in most of the cases oral cancer is diagnosed at last stages or advanced stages where treatment or recovery is almost impossible. However, reports show that the survival rates of oral cancer patients can be increased if it is detected in early stages. Medical experts now a days use various traditional and advanced technologies to identify or detect malignancies from medical images. Similar to the methodology used by medical experts, machine learning methods have been used by many researchers to detect malignancies from medical images. In recent past a lot of research has been done on artificial intelligence and machine learning and a lot of problems have been identified where we can apply these techniques to get a better solution. To name a few areas in [2-4] first time use of machine learning has been reported in websites and social platforms for predicting certain online trends. In [5] machine learning has been used in agriculture science for prediction of crop yield with the help of soil analysis. One area which has taken the interest of most of researchers is analysis of medical disorders and their early detection using machine learning techniques e.g., in [6] authors used machine learning for segmentation of cervical cells from pap smear images for screening of cervical cancer in early stages. In [7] very interesting research on detection of Alzheimer’s disease from CT scans has been done using a combination of neural networks and fuzzy techniques. In [8], a light-weight practical framework for fecal detection and trait recognition is proposed. Author proposed a threshold-based segmentation scheme on the selected color. The images are categorized into five classes with a light-weight shallow CNN. In [9], authors have proposed a CNN model named StoolNet which is capable of detecting and segmentation of stool images. After segmentation classification of stool color is also done with very good accuracy. In [10] authors have used CNN for detection of oral squamous carcinoma from histopathology images with very good accuracies as compared to expert pathologist. However, most of the researchers use traditional machine learning methods that heavily depend on the hand-crafted feature extraction which is complex and time-consuming process. However, with the advent of deep learning this problem is solved due to the automatic feature extraction from the medical images. In [11] authors have combined traditional machine learning methods and deep learning techniques to create hybrid feature extractor that can enhance the classification accuracies of histopathology images. In this paper, we propose a novel yet simple channel based Convolutional neural network (CNN) that will extract the feature from different colour channels and their combinations. We hypothesize that this channel-based approach will help in better detection and classification of oral cancer from histopathology images. To authenticate the proposed methodology, we also investigated the performance of our method on publicly available datasets like BreakHis [12] and ICML [13]. We also investigated our proposed method for HSV and YCbCr color spaces and the effect of individual color channels on classification accuracy.
II. MATERIALS AND METHODS
The dataset that has been used in this research work is collected from Indira Gandhi Govt. Dental College and Hospital, Jammu. Dataset consists of well labeled digital histopathology images of 137 histopathology slides collected from 62 patients of oral carcinoma. These digital histopathology images were captured with the help of camera fitted microscope Nikon NIS F 3.2 at magnification of 400X. These captured images were very large in size (500 to 700 MB) and dimensions (5000x5000). For preprocessing stain normalization method proposed by Macenko et al., in [14] has been used to enhance the histopathology image quality. Macenko et al proposed that instead of normalization in RGB color space the optical density space is more robust. Author transformed RGB images into optical density space and found a threshold empirically for each color channel and normalized the remaining color after thresholding. Author reported that this method of stain normalization proved to be very efficient than other conventional stain normalization method. The method proposed by author in his research paper has been almost used by every researcher that is working on similar research problems. Stain normalization has been used by researchers working on similar works and is commonly reported that it is very helpful for increasing detection rate of histopathological images. To filter out irrelevant or inconsistent images and remove the unwanted part from images we have divided these images into patches of size 300x300 pixels because the input size of Efficient Net is of same dimension. After patch extraction we have a total of 5,718 well labeled images, out of which 2283 are normal tissue images and 2908 are cancerous or having variable degree of carcinoma. 527 patches were background tissue and inconsistent images hence these 527 images were rejected by expert during labelling. Since these images are not sufficient for training and testing of deep learning models so we did data augmentation on these images. For data augmentation we used geometric transformation operations like rotation at 90, 180 and 270 degrees followed by flipping of images to enrich the dataset with augmented images. After pre-processing and data augmentation step, our dataset contains a total of 31,146 histopathology patch images out of which 13,698 images are normal and 17,448 are carcinoma images.
In recent years CNN has been extensively used by researchers in solving challenges like classification of medical imaging data and computer vision. A recent development in deep learning models is Efficient Net which is better than previous CNN both in terms of accuracy and efficiency because it requires less parameters and FLOPS (Floating point operations Per Second). Efficient Net are a family of CNN that consist of 7 CNN named b1 to b7 with b0 as baseline network inspired from existing Mnas Net [15]. Efficient Net is estimated at an average 8.4x smaller and 6.1X faster than best available CNN available because it uses compound scaling [16] and mobile inverted bottleneck convolution blocks (MBconv) [17] along with squeeze and excitation optimization [18]. Compound scaling is the solution to the problem of scaling of different aspects of CNN in a uniform manner, compound scaling uses a compound scaling coefficient ‘Ø’ that is used to scale our CNN uniformly in terms of network width, network depth and resolution. Equation 1 is used to relate depth, width and resolution of a CNN.
such that α.β^2.ϒ^2≈2 and α≥1, β≥1, γ≥1.
where α, β, γ are constants whose values can be found by a small grid search. Intuitively, φ is a user-specified coefficient that controls how many more resources are available for model scaling, while α, β, γ specify how to assign these extra resources to network width, depth, and resolution respectively. Empirically, it can be showed that, compound scaling follows a constraint: α·β2 ·γ2 ≈ 2. By finding different values for Ø we can scale up our baseline network B0 from B1 to B7. Optimal values for B0 network are α = 1.2, β = 1.1, γ = 1.15 as derived from equation (1). It has been shown by authors in [16] that compound scaling is beneficial than scaling only one dimension for overall improvement of CNN.
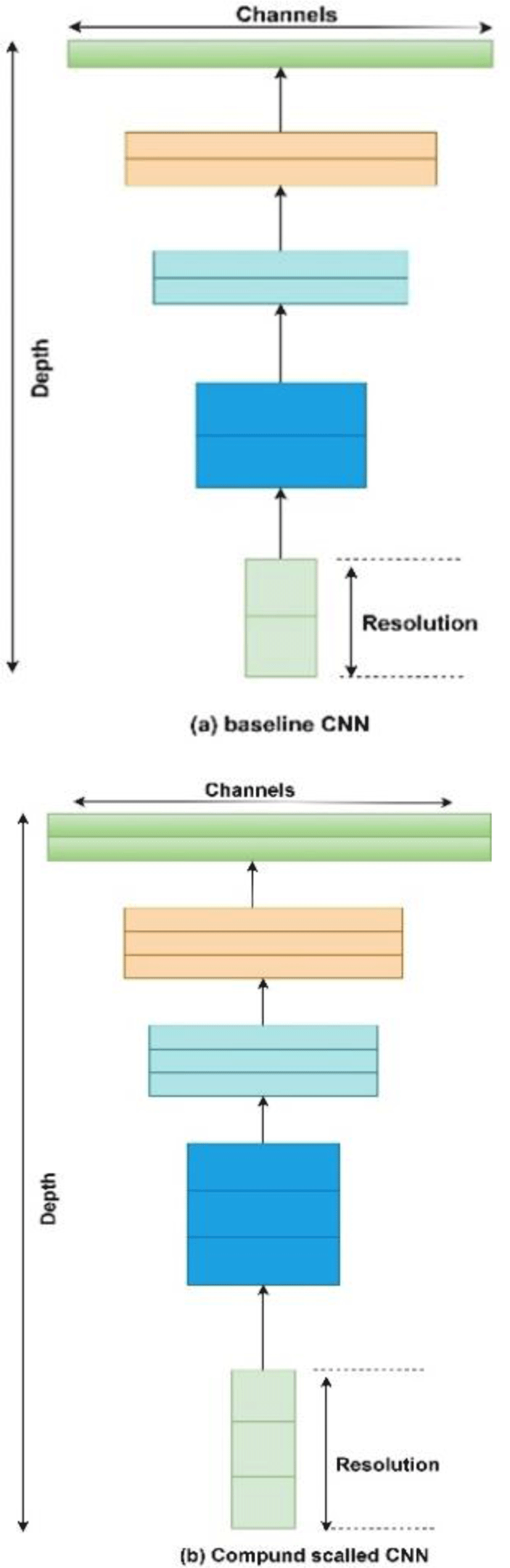
Table 1 represents the architecture of Efficient Net B3 used in this research work. Column 1 shows the number of blocks in our network represented by ‘i’. column 2 shows the name and kernel size of the convolution layers in corresponding block. Column 3 represents the change in resolution of image in different layers. Last column shows the total number of layers in each block.
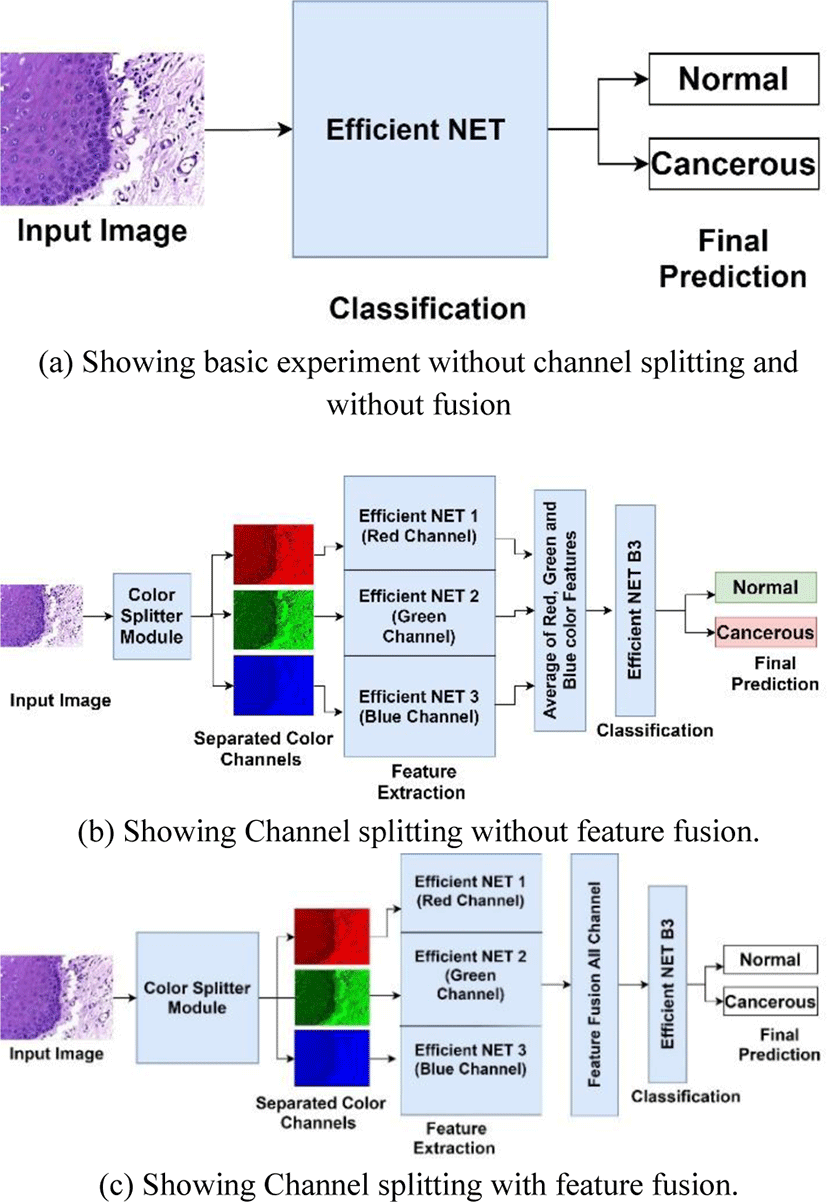
Feature fusion is a technique in which combination of features from different layers or branches of a CNN is done. Feature fusion ensures that the fused features provide a mix of low level as well as high level features from various layers of network. Fusion of features helps in enhancing the classification accuracy of CNN due to its diversity in features as well as all the combination of high level and low-level features. Feature fusion is often implemented via simple operations, such as averaging, summation or concatenation. In this research article we will use averaging and concatenation of three feature vectors from red, blue and green color channels respectively to form a single feature vector that will be used for classification. Fig. 2(a), 2(b) and 2(c) show overall methodology of our research work done in this article.
In this research work we have done the classification of histopathology images of oral cavity into normal and cancerous images. The research methodology followed in carrying out this research work is shown in Fig. 2. All the experiments were performed on intel i5-8265U @1.6 GHz CPU with 8 cores and 24 GB RAM and Nvidia GeForce MX130 GPU. The experiments were done in Python environment on Windows 10 with support of Open CV 20.0.
Experiments were conducted in 2 major phases initial phase was to investigate the classification capability of Efficient Net B3 on oral histopathology images without color channel separation and without feature fusion as shown in Fig. 2(a). For this experiment, mini batch size was initialized as 8, max epochs were set at 20 and learning rate was initialized at 0.0001. Efficient Net B3 was trained using stochastic gradient descent with momentum at 0.9. For performance evaluation purpose accuracy, sensitivity and specificity are used. The dataset was divided into training, validation and testing sets in a ratio of 70:10:20 respectively. This splitting ratio of dataset is kept common for all the experiments conducted in this research paper for fair evaluation of performance.
In second phase 2 experiments was carried out to investigate the performance of Efficient NET when features are extracted from three colour channels red, green and blue respectively with feature fusion and without feature fusion as shown in Fig. 2(b). In this experiment the used averaging of three feature vectors from 3 channels to create a final feature vector for classification. In 2nd experiment the histopathology images were converted into red, green and blue channels only by using channel filter toolbox in OpenCV module. These 3 color channels images were fed parallelly to 3 same Efficient NET model for feature extraction as shown in Fig. 2(c). These 3 efficient Nets are designed in a way that they share same parameters, same input size, same number of layers and kernels, and same size of output. These Efficient Nets were modified by omitting its fully connected and out layers so that it can only extract features. When feature vectors were obtained from all 3 Efficient Nets then feature fusion was done by simply concatenation into one feature vector. This feature vector was fed into the Efficient NET B3 with modified fully connected layers according to size of fused feature vector. The original size of first fully connected layer of Efficient Net B3 is 1536 so when we did feature fusion the modified input size was changes to 1536x3. Initial layers of Efficient NET B3 which was used for classification purpose only were frozen so that fused feature will be directly fed to its input layer. All Efficient NET were configured with same parameters and same values as in experiment 1 for fair comparison.
Table 2 shows the results for the experiments we have done, one can clearly see that classification accuracy in second experiment has increased by 5.73%, sensitivity and specificity also increased by 4.59% and 6.20% respectively. In case of 3rd experiment, the accuracy increased by 1.1% which indicates that channel filtering has some role in feature extraction but since we averaged out the feature vectors so this increase is almost same as first result. However, specificity decreased by 2% from initial result and sensitivity increased by 1.03% which is not significantly different from results reported from first experiment.
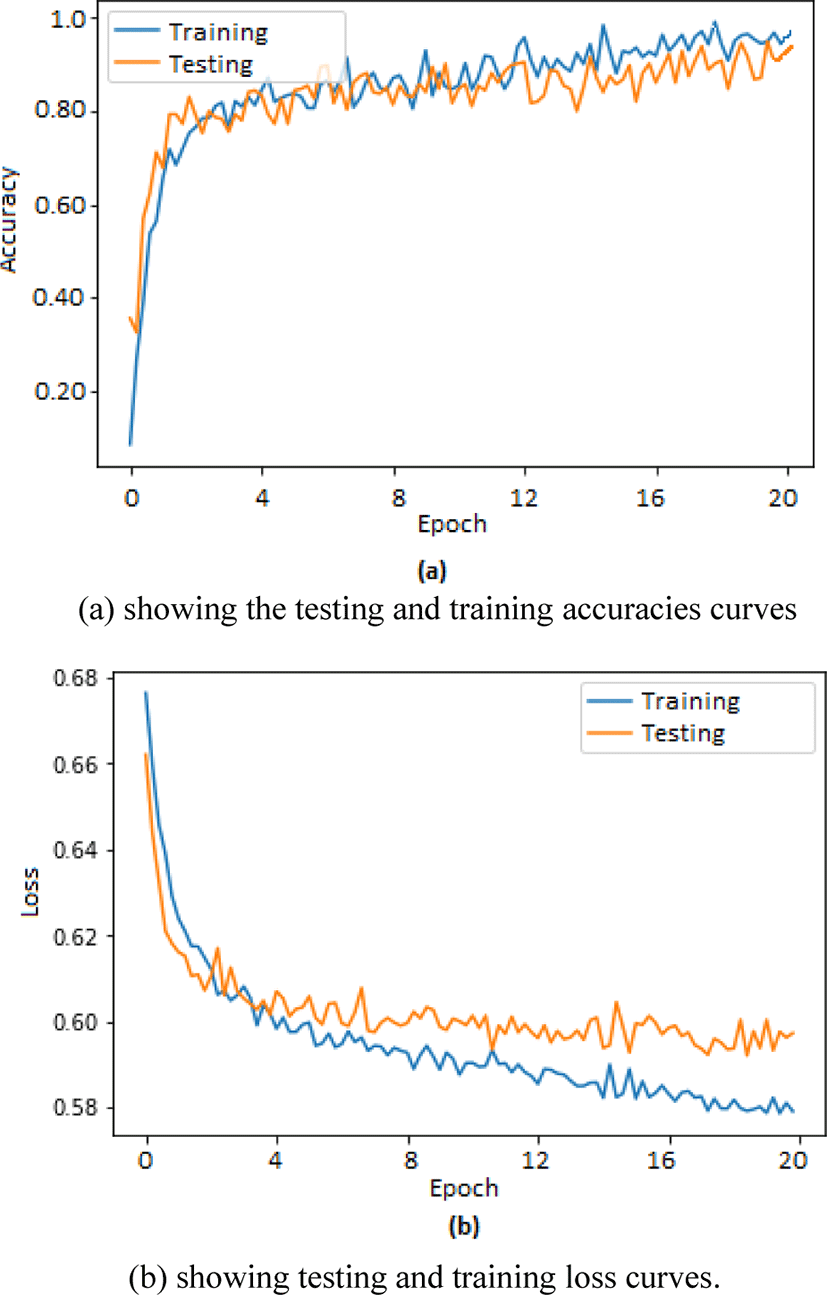
Two separate graphs were plotted using matplotlib toolbox in python for depicting training and testing accuracies in one graph whereas loss in training and testing is depicted in second graph. The graphs shown in Fig. 3(a) corresponds to the training and testing accuracies plotted for 20 epochs whereas graphs shown in Fig. 3(b) corresponds to the training and testing loss recorded during training and testing of our proposed method.
To evaluate the performance of our proposed method we also performed similar experiment on two publicly available datasets BreakHis [4] and ICML [5] datasets, both these datasets are available on Kaggle for public use under open License CC0: Public Domain. BreakHis data set is histopathology dataset containing 588 benign and 1232 malignant histopathology images at 400x magnification. BreakHis also has images at 10x and 40x but we used only 400x images for our experimentation. ICML dataset is also a breast histopathology image dataset containing 28640 patches of histopathology out of which 19861 are normal images and 8879 are cancerous images. We have taken a balanced dataset from ICML by randomly selecting half from normal images. Hence the ICML data that we used for testing our method contains 9930 normal images and 8879 cancerous images. ICML dataset has images of size 50x50 which is a bit smaller hence we resized these images using OpenCV resize method to 300x300. This process compromised the quality of images but Efficient NET accepts only 300x300 input size so we have to resize the images to the input size. These datasets are also stain normalized before splitting the color channels. All parameters were kept same as for our datasets including training, validation and testing size. Table 3 shows a comparison of results achieved by other researchers and our proposed method.
| Dataset used | Author | Method Used | Accuracy |
|---|---|---|---|
| BreakHis | Spanhol et al. [12] | Artificial Neural Network | 83.3% |
| Quadratic Discrimination Analysis | 87.9% | ||
| SVM | 86.1% | ||
| Random Forest | 84.8% | ||
| BreakHis | Deniz et al. [19] | Transfer Learning | 92.04% |
| BreakHis | Spanhol et al. [20] | CNN | 92.03% |
| ICML | Janowczyk et al. [21] | CNN | 76.5% |
| ICML | Angel et al. [22] | CNN | 84.23% |
| BreakHis | Our proposed model | Efficient Net B3 | 94.10% |
| ICML | Our proposed model | Efficient Net B3 | 82.9% |
It can be clearly seen from the Table 3 that our method performed very well on BreakHis dataset and achieved 94.10% classification accuracy whereas in case of ICML dataset our proposed method performed good and was able to achieve 82.9% accuracy. The reason for this performance difference is that the BreakHis dataset we have used contains histopathology images captured at 400x magnification power which is exactly same as our dataset so our model performed very well on BreakHis. In case of ICML the histopathology images are 50x50 in size which is not an optimal size in case of Efficient Net because it needs input of 300x300 size so it may be the major reason that our proposed model did not achieve high accuracy rate as compared to BreakHis dataset.
Apart from RGB color space we also investigated the performance of our proposed model in other color spaces like hue saturation value (HSV), YCbCr where Y is luma part of the image features, Cb is the blue difference of luma and Cr is the red difference of luma. We investigated that if our model works similar as it worked on RGB images. We performed 2 experiments to find out, the results of these two experiments are recorded in Table 4.
| Color space | Accuracy | Specificity | Sensitivity |
|---|---|---|---|
| HSV | 90.2% | 88.73% | 90.39% |
| YCbCr | 85.60% | 84.9% | 86.40% |
From Table 4 it is evident that for HSV color space our model performed very good but for YCbCr color space it performed decent. The reason for this decrease in performance may be the non-linear encoded light intensity in YCbCr color space which is absent in RGB and HSV color spaces. However, we also noticed that HSV color space suffers from varied degree of light intensity component so, it needs all the data with light intensity otherwise it will change the color appearance. Our dataset contains samples which may be having different light intensities because data was captured in different laboratories under different lighting conditions. We can overcome this issue by scaling the light intensity level of all images to same level but it is very computational expensive task and may interfere with natural color of the image. Which is not a very good thing as color is considered a very important feature in diagnosis of cancer in histopathology images.
Findings of above experiments made us curious about what is the impact of individual color in classification problem. So, we investigated the impact of each individual color channel on the classification accuracy. We used only one channel to extract features and suppressing the information from other two channels. This method was repeated for all three individual channels. The results of this experiment are recoded in Table 5.
| Color Channel | Accuracy |
|---|---|
| Red Channel | 62.48% |
| Green Channel | 43.92% |
| Blue Channel | 65.37% |
The results showed that red and blue channels have more information encoded in them than the green channel. However, no single channel can be used for feature extraction alone as no channel has complete information regarding the full image. Combination of all channels gives better results as already stated in Table 2.
III. DISCUSSION AND CONCLUSION
In this research work we have proposed a deep learning-based model for efficient detection of cancerous tissue from histopathology images. Primarily this work is focused on the classification of cancerous tissue images from oral cavity but experiments on other histopathology datasets like BreakHis and ICML which are histopathology datasets related to breast suggest that our proposed method can be used efficiently on other histopathology images also. Obviously when we consider using this model on other histopathology datasets, we have to make some fundamental changes in our model like changing input size, and other hyperparameters accordingly. Our model used red, green and blue channels individually to extract features from image rather than using one single combined RGB channel. Due to separate channel feature extraction the network was able to extract better feature and hence it was able to perform better in classification of images. Feature fusion also played a very important role in enhancing the classification performance as can be seen from the results in Table 2. This work produced very good results but however there are limitations in this work also. Feature fusion done in this work is based on uniformity of channels and filters used in different layers of network. If we wish to concatenate features from different size images or different sized filters the performance of worsened and it may not classify images with good accuracy. We investigated our proposed approach on HSV and YCbCr color space and found that it performed well on these color spaces but not as good as on RGB color space because of the on uniformity of color intensities. Also, we came across an interesting aspect whether individual channels can be used for full image classification. We investigated the significance of individual color channels and found that no individual channel can be used for classification however in future we are interested to investigate the combination of different channels and their impact on classification. Also, we are working on our future work to device a method that can fuse features which are not uniform by nonuniform we mean features from different layers with different kernel size, different filter size and different size of output vectors.