I. INTRODUCTION
Heart rate variability (HRV) refers to the variation in time between consecutive heart beats. This variation is under the control of autonomic nervous system (ANS), which is divided into sympathetic and parasympathetic branches. There have been numerous studies describing the relations of ANS to HRV. Generally speaking, the sympathetic branch tends to increase heart rate (HR) and decrease HRV, whereas the parasympathetic branch tends to decrease HR and increase HRV [1]. A cardiacally-healthy person tends to have both well-functioning sympathetic and parasympathetic branches. As a result, they tend to have higher variability of HRV than the less cardiacally-healthy person.
Main areas of applications of HRV analysis include the risk stratification of sudden cardiac death after acute myocardial infarction. HRV analysis is also generally accepted to provide an early warning sign of diabetic neuropathy. Besides these main applications, HRV has been studied with relation to several cardiovascular diseases, renal failures, physical exercise, occupational and psychosocial stress, gender, age, drugs, alcohol, smoking, and sleep [2].
A number of studies report that each cardiac-related disease has unique HRV in terms of time-domain parameters, frequency-domain parameters, and nonlinear parameters. These parameters have become the de facto numerical/statistical symptoms for each disease. Due to the repetitive and patterned nature of HRV analysis for diagnosis of various diseases, their automation has become increasingly prevalent in our lives. Linear Discriminant Analysis (LDA) has been developed to detect acute mental stress due to university examination with a total classification accuracy, a sensitivity, and a specificity rate of 90%, 86%, and 95%, respectively [3]. Support Vector Machine (SVM) – Radial Basis Function (RBF) and Neural Network (NN) have been developed to detect arrhythmia with an equal average accuracy, sensitivity and specificity of 98.9% [4].
This paper proposes a method to automatically diagnose various diseases. The input data is raw electrocardiogram (ECG) recordings, and the output is multilabel classification of various diseases. The rest of the paper is organized as follows: in Section 2 we briefly review the time-domain parameters, frequency-domain parameters, and nonlinear parameters of HRV analysis. The proposed method is described in Section 3, while Section 4 contains the implementation and discussion of the proposed method. Section 5 contains the conclusion, and finally, future work is presented in Section 6.
II. BACKGROUND
This section describes briefly the prospective analyses that are done to HRV extracted from ECG recordings. The prospective analyses include time-domain parameter analysis, frequency-domain parameter analysis, and nonlinear parameter analysis. The computations as well as the notations used are mainly based on the guidelines given in [5]. A summary of the analysis parameters is given in Table 1.
| Parameter | Unit | Description | References |
|---|---|---|---|
| Time-domain analyses: statistical methods | |||
| Mean RR | ms | The mean of RR intervals | [5] |
| SDNN | ms | Standard deviation of RR intervals | [5] |
| RMSSD | ms | Square root of the mean squared differences between successive RR intervals | [5] |
| NN50 | count | Number of successive RR interval pairs that differ more than 50 ms | [5] |
| pNN50 | % | NN50 divided by the total number of RR intervals | [5] |
| Time-domain analyses: geometrical methods | |||
| HRV triangular index | - | The integral of the RR interval histogram divided by the height of the histogram | [5] |
| TINN | ms | Baseline width of the RR interval histogram | [5] |
| Frequency-domain analyses | |||
| VLF, LF, HF power | ms2 | Absolute powers of very low frequency band (0-0.04 Hz), low frequency band (0.04-0.15 Hz), and high frequency band (0.15-0.4 Hz), respectively | [5] |
| LF/HF ratio | - | Ratio between LF and HF band power | [5] |
| Nonlinear analyses: Poincaré plot | |||
| SD1, SD2 | ms | Short-term and long-term variability standard deviation, respectively | [9,10] |
| Nonlinear analyses: correlation dimension | |||
| ApEn(0.2), ApEn(rmax), ApEn(rchon) | - | Approximate entropy where the tolerance value r is chosen to be r=0.2×SDNN, in the interval [0.1×SDNN, 0.9×SDNN] which maximizes ApEn, and computed according to the following formula proposed by Chon[12], respectively | [11,12] |
| SampEn | - | Sample entropy | [11] |
| Nonlinear analyses: correlation dimension | |||
| D2 | - | Correlation dimension | [13] |
| Nonlinear analyses: detrended fluctuation analyses | |||
| α1, α2 | - | Short-term and long-term fluctuations, respectively | [14.15] |
| Nonlinear analyses: recurrence plot | |||
| lmean | beats | Mean line length of diagonal lines | [16-18] |
| lmax | beats | Maximum line length of diagonal lines | [16-18] |
| REC | % | Recurrence rate (percentage of recurrence points) | [16-18] |
| DET | % | Determinism (percentage of recurrence points which form diagonal lines) | [16-18] |
| ShEn | - | Shannon entropy of diagonal line lengths’ probability distribution | [16-18] |
Time-domain analyses are computationally simple and they are applied directly to the series of successive RR interval values. Also, they do not require stationarity in the same manner as most frequency domain and nonlinear analyses do. There are two methods in time-domain analysis: statistical methods and geometric methods. Statistical methods are based on various moments of the RR intervals and the delta RR intervals. Geometric methods convert the RR interval data into a geometric pattern. The geometric techniques generally have better performance on poorly edited data [6]. The main limitation of time domain analyses is their lack of discrimination between effects of the sympathetic and parasympathetic autonomic branches [7].
The main idea behind the frequency-domain analyses of HRV is the observation that HRV is composed of certain well-defined rhythms, which are related to different regulatory mechanisms of cardiovascular control [7]. Frequency-domain analyses consist of first calculating the power spectral density (PSD) of the RR intervals. Secondly, the PSD is broken into separate frequency bands: very low frequency (0-0.04 Hz), low frequency (0.04-0.15 Hz), and high frequency (0.15-0.4 Hz). Thirdly, the power in each band is calculated by integrating the PSD within the band limits [6]. It is believed that the power in the low frequency band associates with the combination of sympathetic and parasympathetic autonomic branches, while the power in the high frequency band only associates with parasympathetic autonomic branch. Because the high frequency component of HRV is centered around respiratory frequency, respiration should always be considered in HRV analysis [2]. The respiratory frequency can be estimated from the ECG signals – more precisely, from the R-wave amplitudes [8].
Nonlinear analyses of HRV are employed because linear analyses such as time- and frequency-domain analyses are insufficient to describe the complexity of the heart. Therefore, various nonlinear analyses are applied to HRV to fully capture the characteristics of beat-to-beat variability. Nonlinear properties of HRV were analyzed by the following methods: Poincaré plot [9,10], approximate and sample entropy [11,12], correlation dimension [13], detrended fluctuation analysis [14,15], and recurrence plot [16-18]. It is important to note that nonlinear analyses tend to reveal more information about HRV characteristics than time- or frequency-domain analyses.
III. PROPOSED METHOD
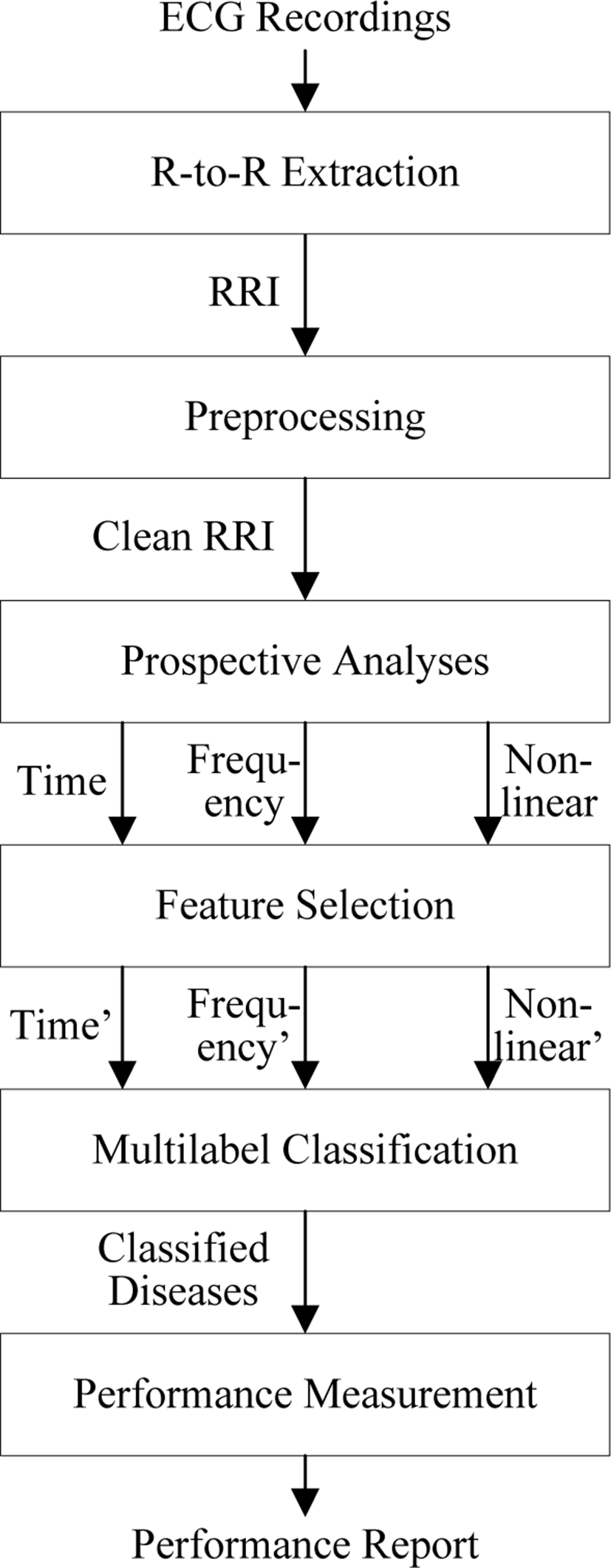
The overall procedures for automated diagnosis of various diseases are shown in Figure 1. The input data is raw
ECG recordings, from which the R-to-R intervals (RRI) of HRV are extracted. The requirements of the RRI signal to be processed are freedom from ectopic beats (abnormal beats that are due to unusual impulses), stationarity (absent of low frequency trends), and evenly-sampled RRI [19]. To achieve these three requirements, the RRI signal is initially preprocessed.
After that, prospective analyses are performed on the preprocessed RRI signal to extract time-domain parameters, frequency-domain parameters, and nonlinear parameters of RRI signal, as explained in Section 2. Those parameters are unique for each disease and can be used as the statistical symptoms for each disease. It is possible to utilize all HRV parameters reported in Table 1 for performing diagnosis on various diseases, however this may decrease the performance of the classifier, particularly because of the curse of dimensionality [20]. Therefore, feature selection is performed to obtain only the essentials of time-domain, frequency-domain, and nonlinear parameters.
The selected features (i.e. selected parameters) are then utilized to diagnose various diseases using machine learning techniques. Machine learning techniques suitable for this task are multilabel classification techniques, such as Artificial Neural Networks (ANN), random forest, Support Vector Machine (SVM), and Linear Discriminant Analysis (LDA). In order to evaluate the classifier, common measures are computed for binary classification performance measurement [21] for each class. The classification performance is described in terms of statistical accuracy, sensitivity, and specificity.
The details of the proposed method are summarized in Table 2.
IV. IMPLEMENTATION AND DISCUSSION
The proposed method above is implemented inside MATLAB® R2016a environment (The MathWorks, Inc.). The proposed method is still a work in progress; therefore, not all of the procedures have been implemented. The procedures that have been implemented are R-to-R extraction and preprocessing.
The input data used to test the system is the standard MIT-BIH arrhythmia database. A record consists of three files; the header file, the annotation file, and the data file. The header file is a short text file that describes the signals, including the name or URL of the signal file, storage format, number and type of signals, sampling frequency, calibration data, digitizer characteristics, record duration, and starting time. The annotation file contains sets of labels, each of which describes a feature of one or more signals at a specified time in the record. The data file contains digitized samples of one or more signals.
By nature, the raw ECG data is non-stationary; that is, there are low-frequency trends in raw ECG data. Therefore, the first step is to remove the low-frequency component [22]. This process is done by applying Fast Fourier Transform (FFT), removing low-frequencies, and restoring the ECG signal by applying Inverse FFT (IFFT).
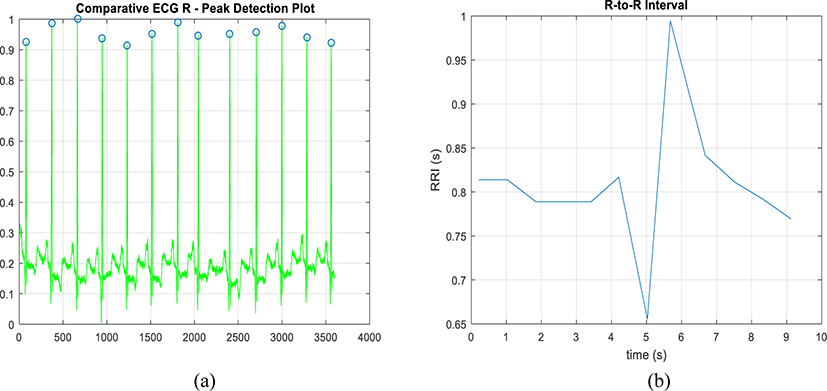
The second step is to find the local maxima. To do that, apply windowed filter which detects the maximum in its window only and ignores all other values. Only the significant values of the windowed signal should be preserved. To do this, use threshold filter. In order to refine the result and ensure that all the peaks are detected, adjust the filter window size and repeat filtering.
Using MITDB 100 data, the R-peaks detection and extraction works as expected. The result for R-peaks detection and R-to-R extraction for the first 10 seconds is shown in Figure 2.
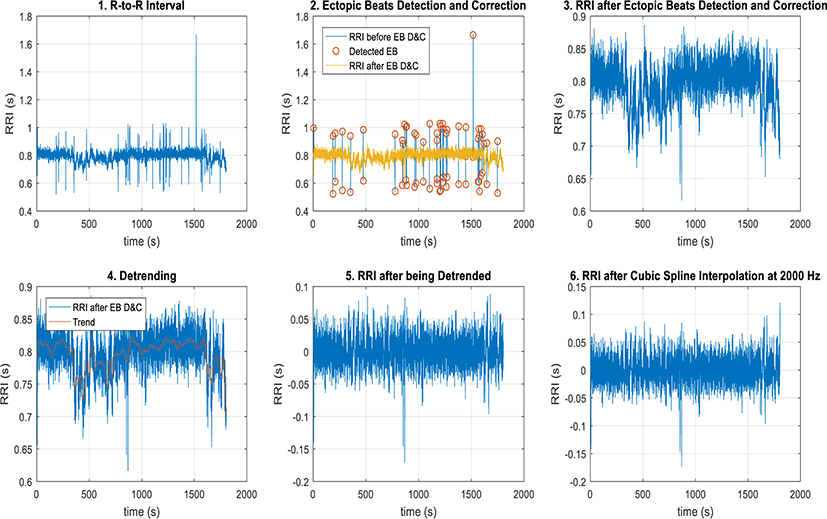
In preprocessing, the signal is required to have no ectopic beats, to be stationary, and to be evenly-sampled. Therefore, there are three subprocedures that needs to be done in this step. In order to have the signal with no ectopic beats, two methods are used: ectopic beats detection based on thresholding and ectopic beats correction based on linear interpolation. In order to have stationary signal, the method of detrending based on smoothness prior approach is used.
In order to have an evenly-sampled signal, the method of resampling based on cubic sampling interpolation is used.
Using MITDB 100 data, the preprocessing method works as expected. The step-by-step procedure of preprocessing is shown in Figure 3.
V. CONCLUSION
Due to the repetitive and patterned nature of HRV analysis for the diagnosis of various diseases, their automation has become increasingly prevalent in our lives. In this paper, a new method was proposed for automated diagnosis of various diseases, as explained in Section 3. The first two steps, which are R-to-R extraction and preprocessing, have been successfully implemented with satisfactory results.
VI. FUTURE WORK
There is still a need to explore in detail various alternatives for statistical analysis methods, feature selection methods, multilabel classification methods, and performance measurement methods. After that, these methods would need to be integrated and implemented to form one large system capable of automated diagnosis of various diseases.