I. INTRODUCTION
Alzheimer’s disease (AD) which mostly affects older people is a common neurodegenerative brain disease. Mild Cognitive Impairment (MCI) is a phase in which a person has mild but noticeable changes in thinking patterns. Although there is no medication to cure Alzheimer’s, some medications have been practiced delaying the onset of memory-related symptoms on the patients [1]. Due to this reason, proper diagnosis of AD and MCI in the initial phase plays a crucial role. AD is a frequently occurring neurodegenerative disorder influencing many individuals worldwide. Finding specific and sensitive biomarkers on the early Alzheimer’s progression is crucial to help clinicians and researchers to initiate new diagnostic and treatments capability, in addition, to shortening the cost burden and time of clinical examinations. To date, the identification of Alzheimer’s is mainly relying on clinical observation and neuropsychological evaluation [2]. In the year 1980s, the National Institute of Neurologic and Communication Disorders and Stroke and the Alzheimer’s disease and Related Disorders Association (NINCDS-ADRDA) established clinical diagnostic standards for Alzheimer’s based on binary technique. Mentioning in this method, cognitive damage is fundamental for the identification of Alzheimer’s [3]. Neuropathological evidence built on neurofibrillary tangles and senile plaques were introduced later [4].
In the year 2011, National Institute on Aging-Alzheimer’s Association Group improved the diagnostic regulation for Alzheimer’s. These improved diagnostic regulations have occupied the binary technique for a further pathological explanation of disease: supplementary features can be received by measuring Cerebrospinal Fluid (CSF), neurogenetic approach, tau, amyloid, and neuronal damage features as assessed by neuroimaging analysis, including Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET) and Functional Magnetic Resonance Imaging (fMRI). MRI and PET imaging changes provide a measurement of atrophic regions and amyloid/metabolism biomarkers [5, 6], to find AD, even at an early stage of Alzheimer’s apparent [7]. Due to the MRI modality’s non-invasiveness, a substantial amount of effort has been set into an improvement of the MRI processing scheme to find MRI-associated features, that can be utilized for boosting the efficiency of Alzheimer’s diagnosis. Major investigation, which was concentrated on the identification of MR image variations between an individual with normal and AD subjects. MRI technology provides a highly reliable and intuitive basis for the identification of disease. Most of the research has proved that specific anatomical parts of AD and MCI individuals are sensitive to neurodegenerative progress. In the initial phase, individuals undergo functional neuronal damage due to loss of gray matter and change in cortical thinning [8]. Several studies have noted that with the advancement of the pathology, there is critical atrophy has been noticed in the medial temporal region, such as the amygdala, hippocampus, para-hippocampal cortices, and entorhinal parts. Therefore, structural MRI will thus have an extensive role in the identification of AD, and it will have a great role in medication. The progress of powerful, automatic, significant techniques to utilize MRI brain images is therefore crucial stance to further widen the utility of structural imaging in the situation of neurodegenerative diseases. A huge number of articles have pointed out the correlations among quantitative markers from MRI with the progression of AD. The performance of automatic detection is related to trained export to disease identification using MRI images of AD individuals.
There is large documentation that indicated that various anatomical brain parts are affected at various phases of the pathology, with initial involvement of the amygdala, hippocampus, and entorhinal cortex. Even though these parts are responsible for Alzheimer’s, these regions have yet to analyze carefully. Common methods that take a pathological variation of multiple parts within the entire brain consider improving the accuracy in Alzheimer’s analysis and to assist the differential assessment of various kinds of brain diseases. It is however considered to follow holistic steps and to analyze whole structures of the entire brain instead of a particular section of the brain. Due to initial involvements of these parts of the brain in Alzheimer’s, the target of several published articles relies on the hippocampal sub-regions to evaluate its shape or volume. Cortical thickness and gray matter tissue features have also been considered highly anticipating measures in the context of Alzheimer’s diagnosis. Other several methods rely on deformation-based analysis; voxel-based analysis; or tensor-based analysis to measure group differences. Commonly, entire brain analyses are effective as compared to individual parts such as the hippocampal regions. It is widely acknowledged that the hippocampus sub-regions play a major role in the short-to long-term memory assimilation process. The hippocampal area is more prone to be the early brain part to endure suffering. Moreover, clinical analysis has shown that the hippocampal region is one of the widely used and most potent biomarkers for recognizing the transformation from MCI to AD [9], [10]. Nevertheless, it is generally considered as a single entity due to the crude resolution of MRI. With the significant progress in high-resolution MRI images data procurement systems, new freedom for especially investigating individual hippocampus sub-regions have developed. It is now feasible to analyze the presubiculum, subiculum, fimbria, hippocampal tail, Dentate Gyrus (DG), Hippocampus-Amygdala-Transition-Area (HATA), hippocampus fissure, hippocampus, and the four Cornus Ammonis regions (CA1-CA4) [11]. It has been proclaimed that CA1 volume measurements were hypersensitive than whole hippocampus volumetry for finding anatomical changes at the early stage of Alzheimer’s [11]. It was also noted that hippocampus sub-regions were linked with age-associated memory loss and specific aspects of memory pattern [11]. The cerebral cortical region reflects the distinct atrophic mechanisms of an individual with AD and MCI. For example, the cortical thickness exposes the rate of brain degeneration by identifying the shortest measures between an outer and inner area of the cerebral cortex. The surface regions denote the measures of cortical pattern, and the difference in mean curvature is produced by variation of surface regions that gives practical clues on the volume changes and folding mechanism in the cerebral cortex. The appropriate properties of cortical regions may reveal different progression trajectories for various neurodegenerative patterns [12]. These measurements show a major role in the diagnostic classification of AD and MCI, and in the initial detection of MCI individuals who carry the risk of onset development of AD [13].
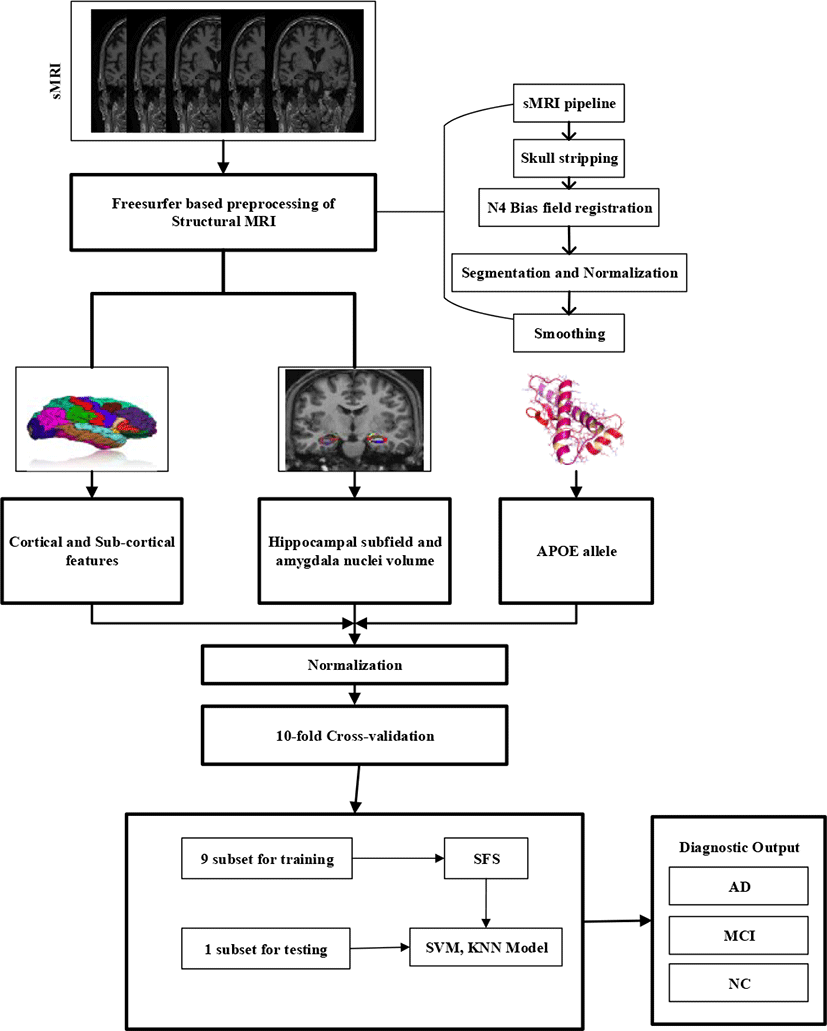
Cortical and subcortical, hippocampal, and amygdala volume changes are considered the major hallmark of AD and therefore it is used as a diagnostic marker. Hippocampal and amygdala atrophy in Alzheimer’s patients usually extend to other brain regions [14, 15]. The pattern of hippocampal amygdala, cortical and subcortical atrophy pattern can be precisely visualized with anatomical MR imaging. Which plays a major role in the clinical detection of AD [16, 17]. Moreover, alteration on cortical thickness [18], as well as widespread cortical and subcortical atrophy have been demonstrated in a patient with AD in comparison to healthy control [19]. In this article, we proposed a classification framework to precisely diagnose an individual with AD and MCI from Healthy Controls (HC). Firstly, we perform the cortical and subcortical segmentation using Freesurfer (v6.0.0) [20], hippocampal subfield, and amygdala nuclei volume segmentation were obtained using segmentHA_T1.sh function available on Freesurfer [20] and genetics data were obtained from the ADNI biomarkers core laboratory. After that, we combined all these measures into the predictive model and calculate the performance of classification. We hypothesize that the combination of the feature will outperform the individual separate model. Fig. 1 gives the overall outline of the suggested method. The rest of the flow of the proposed method is as follows.
II. MATERIAL AND METHOD
The datasets used to develop these studies were acquired through the Alzheimer’s disease Neuroimaging Initiative (ADNI) data bank. In this paper, 211 individuals are used including 54 AD patients, 99 MCI subjects, and 58 normal controls. Table 1 presents the demographic details of all these subjects. All structural MRI scans tested in this paper were obtained from 1.5 T.
MR images used in these studies were obtained through 1.5T scanners. We collect all MRI images in the Neuroimaging Informatics Technology Initiative (NifTi) format. Downloaded images were pre-processed for spatial distortion and B1 field inhomogeneity correction.
The ADNI data bank core laboratory was also granted with gene expression and genotype data for an individual patient in this article, that was retrieved from peripheral blood inspection. The genetic biomarkers were a single definitive variable for every single individual, considering one of five potential measures :( ε2, ε3), (ε2, ε4), (ε3, ε3), (ε3, ε4), or (ε4, ε4). In this article, we particularly investigate APoE ε4 allele aspect (noncarrier vs. carrier).
The prime aim of features selection is to choose several features from the pool of obtained feature vectors that boost the performance accuracy. In this work, we utilized a Sequential Features Selection (SFS) procedure. SFS method relies on the search technique which begins from empty feature vectors S and repeatedly adds features selected by some evaluation function which increases the performance accuracy by reducing the Mean Square Error (MSE). At each step, the new features are to be induced in the features set. So, the new extended features set should produce maximum classification accuracy as compared with the addition of further features vector. SFS is widely utilized for its simplicity and calculation speed. Many previous kinds of literature proposed and applied the SFS algorithm [21].
In this proposed technique, we applied the Multi-Kernel Support Vector Machine (MK-SVM); Support Vector Machine (SVM) is primarily a binary classifier for effective analysis of both linearly non-isolated and isolated vectors. SVM measures the best hyper-plane that classifies the data having maximal classification margin against support vectors as the same time training phase described, during the testing phase on a new data set.
Classifier makes a judgment on the support of hyper-plane. In this work, we used multiple kernels instead of a single kernel, such as Gaussian, polynomial, and sigmoid kernel which can be utilized together. Various kernels available for SVM are united with a convex estate with weight based on their selective potential. For a given training vector, the kernel was defined as ; here we utilized kernel function of , where βυ represent the weight coefficient. The MK-SVM is represented as:
where α represent a Lagrange multiplier and in this method, we decide the optimal value of βυ by Cross-validation (CV) on the training vectors set utilizing grid search in the range of [0,1], maximum validation accuracy was achieved at β = 0.1.
We perform the classification comparison of the proposed classifier with the K-Nearest Neighbor (KNN) technique. The KNN technique is widely used and the simplest machine learning approach. For the training procedure, a labeled database is fed as an input, and then an unlabeled data set is classified on the support of the label of K data set nearest the unlabeled data point of the neighborhood.
Here, K is the main parameter of the algorithm. Euclidean distances were used to define the instance of the nearest neighbors. Which is defined as:
where xi and yi represent two features’ vectors utilized for performance evaluation on a variable number of neighbors to classify Alzheimer’s patients at different value of K=3,4,5…9. Testing data sets are classified according to the specified K nearest neighbours.
We also carried out the Cross-Validation (CV) techniques on classification dataset. CV is a widely used and popular data resampling technique for evaluating the generalization idea of an estimating design and preventing the over or underfitting of the classifiers. CV is widely utilized in predictive modalities such as classification problems. In such type of issue, a design is adapted with a familiar sample space, which is known as the training data, and a sample of unknown data against that the design is evaluated, that acknowledge as the test vector set.
The purpose is to prepare a testing specimen for the design in the training phase, and then provide the light inside how the appropriate design adopts various independent data spaces. Each step of the CV requires the division of data sample into independent data sets, then investigate on every data sample. After that, the process is validated on other sub-data which are known as testing samples.
To lower the fluctuation, several steps of CV are carried out utilizing various distinct separations, and after those results are taken as average. CV is a robust procedure in the model performance evaluation. The accuracy, specificity, sensitivity, Receiver Operating Characteristic (ROC) curve, and Cohen Kappa index were adopted to verify the results of classification. In this method, we referred to the HC as negative samples AD patients as positive samples.
True-Negative (TN) represents the value of negative pattern which is correctly classified; True-Positive (TP) indicates the number of positive patterns correctly categorize; False-Positive (FP) indicates the value of negative pattern classified as positive; similarly, False-Negative (FN) represents the pattern of positive dataset classified as a negative sample. The accuracy, specificity, precision, and area under the curve are defined as:
Receiver Operating Characteristic (ROC), popularly known as ROC is a curve developed by the plot of false-positive rate versus true-positive rate, which can evaluate the performance efficiency of a binary classifier. The ROC calculates the Area Under the Curve (AUC) which is comparable to the performance of the classifiers.
The cortical regions were designed using 3D MPRAGE images by Freesurfer (http://surfer.nmr.mgh.harvard.edu/) software as shown in Fig.2. The complete surface construction scheme with Freesurfer mentions and tested in previous research [20, 22]. In short, the step cover parcellation of white/Gray matter link tessellation, white matter, folded surface inflation, and automatic topological defects correction in the resulting manifold.
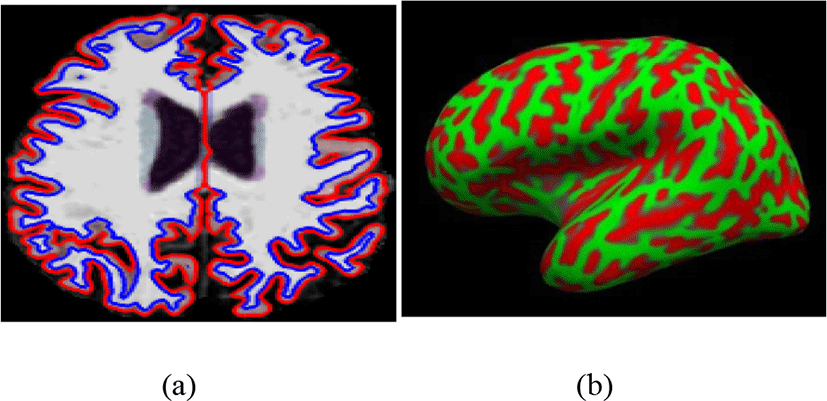
This surface was then used to start for a deformable regions method construction to find the white/Gray and pial regions. Cortical regions thickness for each subject was computed with 1 mm style uniform gird over both cortical regions, thickness is obtained by the precise span between the pial surface and white/Gray matter design [22], implementing in aspect evaluation of submillimetre variation.
The high-resolution surface-based averaging method is used for all images to align a common surface template that regulates cortical regions folding styles. Regions of Interest (ROI) were mapped back on an approved brain to individual’s natural space applying a high-spatial spherical transform to obtain the homologous distribution across subjects. Then mean thickness and Gray matter volume of cortex in individuals ROI were measured. After all, 10-mm half height at full-width Gaussian kernel was used to smooth cortical thickness to decrease local variations for further analysis.
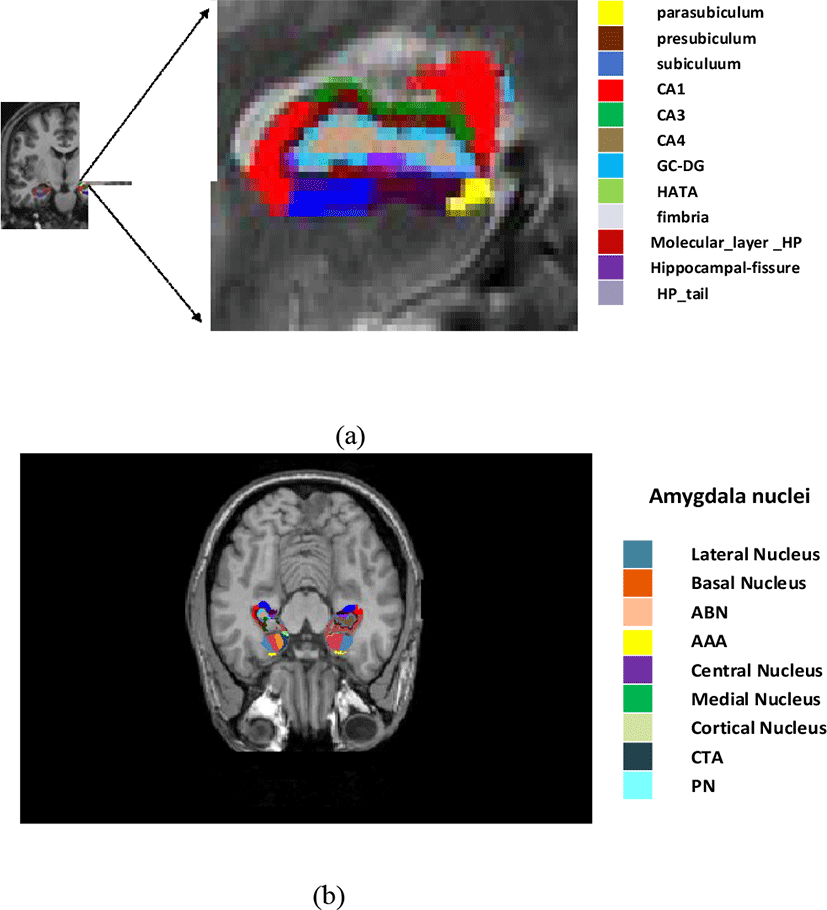
Hippocampal sub-regions in MRI shown to be important in the prediction of Alzheimer’s in the individual having mild symptoms. To calculate atrophy measure on hippocampal even more precisely and predict the Alzheimer’s in MCI as well as individual with normal control it is more important to analyze the sub-volume of hippocampal.
In our method, hippocampal segmentation was performed using Freesurfer [20] tool. Hippocampus atrophy is considered a major indicator of Alzheimer’s disease identification [23]. Hippocampus sub-regions and amygdala nuclei subfields estimation were eventually performed by implementing the hippocampal sub-regions parcellation technique released by Freesurfer (version 6.0.0).
This algorithm employs an atlas-based probabilistic Bayesian interface and is generated with ultra-high resolution ex-vivo MRI imaging data (0.1-0.15 mm isotropic) to create a computerized parcellation of the amygdala and hippocampal regions. Simultaneous segmentation of both structures assures that exclusion of overlap among them and the possibility of a void between these were excluded [24].
Hippocampal parcellation comprised 12 sub-regions: namely subiculum, presubiculum, parasubiculum, cornu amonis fields 1,2/3, and 4 (thereafter indicate to as CA1, CA3, and CA4), granule cell sheet of the Dentate Gyrus (DG), a transition of Hippocampus-Amygdaloid Area (HATA), fimbria (an area of white matter), the molecular coat of DG, fissure region of hippocampal, and the tail of hippocampal.
Amygdala sub-regions are divided into nine regions: accessory basal and basal, central medial, the lateral, cortical, and the anterior amygdala regions, para laminar nucleus, and Cortico-Amygdaloid Transition Area (CTA) as shown in Fig. 3
III. RESULTS AND DISCUSSION
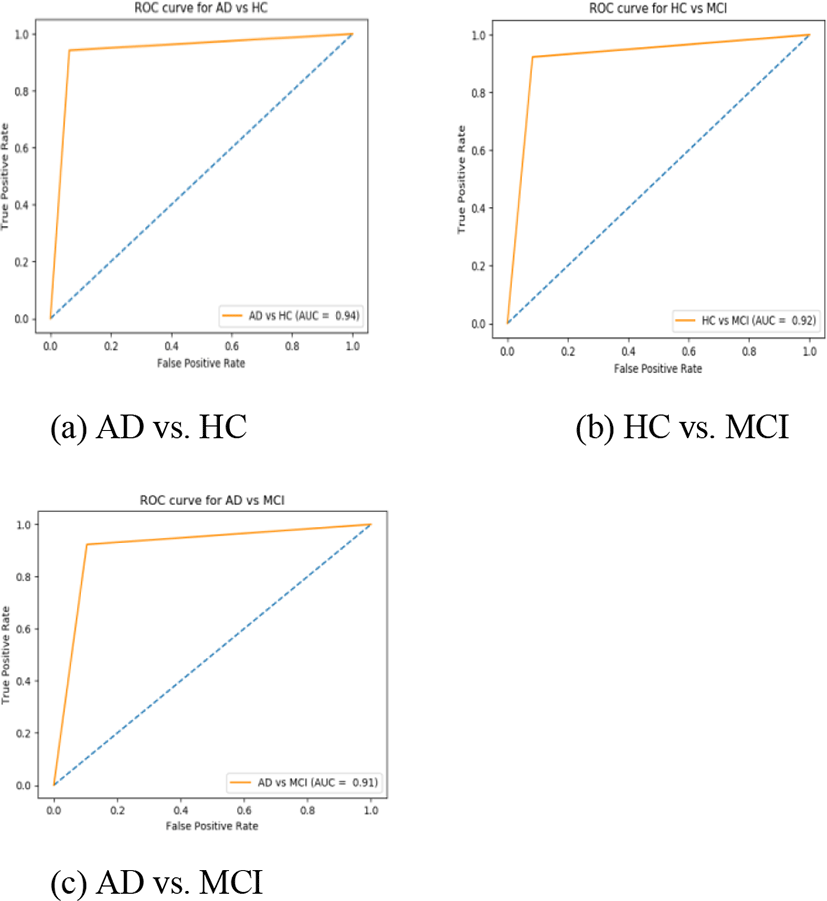
Performance evaluation is measured in classification accuracy, specificity, sensitivity, precision, F1 score, Cohen kappa, and Area Under the Curve (AUC) on receiver operating curve for classification of HC vs. MCI, AD vs. MCI, and AD vs. HC on obtained biomarkers measures individually, and by combining all three features. The accompanying AUC represents the measure of classification accuracy and AUC, as shown in Table 2, 3 and 4 and in Fig. 4 respectively; two reference measure performs reasonably well. Especially, the hippocampal subfield and amygdala nuclei volume features can discriminate well between the AD patients and healthy controls as compared to cortical and subcortical features alone. Most importantly, however, the combination of all biomarkers measures outperforms the separate measure. As results listed in Table 2, 3, and 4 shows that this algorithm gained 94.33% performance accuracy, 95.40% sensitivity, 96.50% specificity, and 94.30% AUC, along with 92.71 % precision, 93.02 % F1 score, and 0.9030% Cohen kappa score on the classification of AD vs. HC. For AD vs. MCI evaluation, our algorithm achieves a performance accuracy of 85.58%, with 95.73% sensitivity, 87.30% specificity, a precision of 87.73%, F1 score of 90.30% with a Cohen kappa score of 0.8735, and AUC of 91.48%. Similarly for classifying HC vs. MCI, our method achieves a performance accuracy of 89.77%, 96.15% sensitivity, 87.35% specificity, 87.38% precision, 85.30% F1 score along with Cohen kappa score of 0.8908, and AUC of 92.55%. To calculate the classification task of our algorithm we adopt sequential features selection (wrapper method) to obtain the influential features set and increase class separability after that we fed that feature set to Multi-Kernel SVM for classification. We also compared the classifier performance with KNN classifiers and obtained the best results. Ten-fold cross-validation is applied for the evaluation of the proposed method. Stratified K-fold CV is used to optimize the hyper-parameter in the classification method.
To inspect the performance of the proposed workflow with the existing state-of-the-art method we utilized ADNI publicly available dataset. The ADNI data bank as the public-private collaboration was established in 2003 with assistance from various associations within the institution that including the National Institute of Aging and National Institute of Biomedical Imaging and Bioengineering, along with various private pharmaceutical companies and non-profit organizations. The fundamental purpose of the ADNI has to observe whether the merging of serial PET, MRI, clinical neuropsychological tests, and biomarkers can assess the conversion of MCI and initial AD syndrome.
In this study, we included 211 sMRI (belonging to HC, MCI, and AD) acquired from the ADNI data bank. All subjects meet with several neuropsychological assessments to measure several clinical characteristics including MMSE score, clinical dementia ratio, and functional assessment questionnaire score. Demographic information about all participants is present in Table 1. Both female and male participants were combined in our analysis. All MR images utilized in this analysis were obtained from an MR scanner with a 1.5T MRI. Initially, we measure the cortical, subcortical, and hippocampal sub-field by applying Freesufer from each sMRI. The proposed study obtained enhanced results for all classification groups (AD vs. MCI, HC vs. MCI, and AD vs. HC). For the AD vs. HC group, the MK-SVM classifier with feature concatenation obtained an AUC of 94.30 % and an accuracy of 94.33 % as compared to the individual features set. Similarly, for the AD vs. MCI group, our method with feature combination achieved an AUC of 91.48% and an accuracy of 85.58%. Furthermore, for HC vs. MCI classification, this algorithm achieved an AUC of 92.55% and an accuracy of 89.77% with combined features set.
Recently, several state–of–arts articles have noted their performance report for identifying Alzheimer’s victims from healthy individuals by utilizing MRI imaging data. Zhang et al. [25] set up a multimodal analysis of AD using features concatenation of MRI, PET, and CSF. Experiment shows that MRI data accomplished an accuracy of 86.2 % while classifying AD vs. HC group. By a combination of all introductory features results improved and they produce an accuracy of 93.2%. Lama et al. [26] utilized the Freesurfer to compute volumetric measures and cortical thickness, using these features set with an extreme learning machine, they obtained an accuracy of 77.88 %. Westman et al. [27] achieved 87% accuracy only with sMRI markers and later increased accuracy to 91% by merging MRI data along with CSF biomarkers features. Performance comparison of these studies among other existing classification algorithms using the ADNI database is listed in Table 5. The below table presents that the classification efficiency of the proposed technique by utilizing features from MR imaging and genetics biomarkers is comparable and superior to that of the existing algorithm listed in the research.
| Method | Modalities | Data | Classifier | ACC | SEN | SPEC |
|---|---|---|---|---|---|---|
| Lama et al. [26] | sMRI | ADNI | ELM | 77.88 | 77.88 | 68.85 |
| Westman et al. [27] | sMRI | ADNI | OPLS | 87 | 83.30 | 90.10 |
| Zhang et al. [25] | sMRI | ADNI | SVM | 86.2 | 86 | 86.3 |
| Proposed method | sMRI+ APoE ε4 | ADNI | MK-SVM | 94.33 | 95.40 | 96.50 |
IV. CONCLUSION
In this paper, first, we extracted the morphometric, cortical-subcortical features, hippocampal subfield, amygdala nuclei volume from sMRI, and genetics features from the ADNI core laboratory biomarkers. By combining these different kinds of features and then using these features set to perform the AD diagnosis, we notice that the combination of all features outperforms as compared to the individual features alone. Besides we used the sequential features selection (SFS) algorithm to obtain the optimum features set of all combined features space, which helps to achieve the maximum classification accuracy. After all, we fed the SFS features into Multi-kernel SVM with a 10-fold cross-validation scheme to achieve the classification result, which validates the effectiveness of the proposed algorithm to improve performance analysis. We also analyzed the performance accuracy with the KNN classifier, which shows the effectiveness of the present algorithm. Furthermore, to improve the potency of this algorithm, we plan to include the longitudinal dataset, increasing the multimodal dataset and different imaging techniques. Ensemble approach and other feature selection methods.